Rolling mean with CMIP6
Contents
Rolling mean with CMIP6¶
Import Python packages¶
import s3fs
import xarray as xr
import intake
import matplotlib.pyplot as plt
import cartopy.crs as ccrs
import numpy as np
Open CMIP6 online catalog¶
cat_url = "https://storage.googleapis.com/cmip6/pangeo-cmip6.json"
col = intake.open_esm_datastore(cat_url)
col
pangeo-cmip6 catalog with 7632 dataset(s) from 517667 asset(s):
| unique | |
|---|---|
| activity_id | 18 |
| institution_id | 36 |
| source_id | 88 |
| experiment_id | 170 |
| member_id | 657 |
| table_id | 37 |
| variable_id | 709 |
| grid_label | 10 |
| zstore | 517667 |
| dcpp_init_year | 60 |
| version | 715 |
Search for data¶
cat = col.search(source_id=['CESM2-WACCM'], experiment_id=['historical'], table_id=['AERmon'], variable_id=['so2'], member_id=['r1i1p1f1'])
cat.df
| activity_id | institution_id | source_id | experiment_id | member_id | table_id | variable_id | grid_label | zstore | dcpp_init_year | version | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | CMIP | NCAR | CESM2-WACCM | historical | r1i1p1f1 | AERmon | so2 | gn | gs://cmip6/CMIP6/CMIP/NCAR/CESM2-WACCM/histori... | NaN | 20190227 |
Create a dictionary from the list of dataset¶
dset_dict = cat.to_dataset_dict(zarr_kwargs={'use_cftime':True})
--> The keys in the returned dictionary of datasets are constructed as follows:
'activity_id.institution_id.source_id.experiment_id.table_id.grid_label'
100.00% [1/1 00:00<00:00]
list(dset_dict.keys())
['CMIP.NCAR.CESM2-WACCM.historical.AERmon.gn']
Open dataset¶
dset = dset_dict['CMIP.NCAR.CESM2-WACCM.historical.AERmon.gn']
dset
<xarray.Dataset>
Dimensions: (lat: 192, lev: 70, lon: 288, member_id: 1, nbnd: 2, time: 1980)
Coordinates:
* lat (lat) float64 -90.0 -89.06 -88.12 -87.17 ... 88.12 89.06 90.0
lat_bnds (lat, nbnd) float32 dask.array<chunksize=(192, 2), meta=np.ndarray>
* lev (lev) float64 -5.96e-06 -9.827e-06 -1.62e-05 ... -976.3 -992.6
lev_bnds (lev, nbnd) float32 dask.array<chunksize=(70, 2), meta=np.ndarray>
* lon (lon) float64 0.0 1.25 2.5 3.75 5.0 ... 355.0 356.2 357.5 358.8
lon_bnds (lon, nbnd) float32 dask.array<chunksize=(288, 2), meta=np.ndarray>
* time (time) object 1850-01-15 12:00:00 ... 2014-12-15 12:00:00
time_bnds (time, nbnd) object dask.array<chunksize=(1980, 2), meta=np.ndarray>
* member_id (member_id) <U8 'r1i1p1f1'
Dimensions without coordinates: nbnd
Data variables:
so2 (member_id, time, lev, lat, lon) float32 dask.array<chunksize=(1, 5, 70, 192, 288), meta=np.ndarray>
Attributes: (12/48)
Conventions: CF-1.7 CMIP-6.2
activity_id: CMIP
branch_method: standard
branch_time_in_child: 674885.0
branch_time_in_parent: 20075.0
case_id: 4
... ...
variable_id: so2
variant_info: CMIP6 CESM2 hindcast (1850-2014) with high-top a...
variant_label: r1i1p1f1
status: 2019-11-05;created;by nhn2@columbia.edu
intake_esm_varname: ['so2']
intake_esm_dataset_key: CMIP.NCAR.CESM2-WACCM.historical.AERmon.gnxarray.Dataset
- lat: 192
- lev: 70
- lon: 288
- member_id: 1
- nbnd: 2
- time: 1980
- lat(lat)float64-90.0 -89.06 -88.12 ... 89.06 90.0
- axis :
- Y
- bounds :
- lat_bnds
- standard_name :
- latitude
- title :
- Latitude
- type :
- double
- units :
- degrees_north
- valid_max :
- 90.0
- valid_min :
- -90.0
array([-90. , -89.057592, -88.115183, -87.172775, -86.230366, -85.287958, -84.34555 , -83.403141, -82.460733, -81.518325, -80.575916, -79.633508, -78.691099, -77.748691, -76.806283, -75.863874, -74.921466, -73.979058, -73.036649, -72.094241, -71.151832, -70.209424, -69.267016, -68.324607, -67.382199, -66.439791, -65.497382, -64.554974, -63.612565, -62.670157, -61.727749, -60.78534 , -59.842932, -58.900524, -57.958115, -57.015707, -56.073298, -55.13089 , -54.188482, -53.246073, -52.303665, -51.361257, -50.418848, -49.47644 , -48.534031, -47.591623, -46.649215, -45.706806, -44.764398, -43.82199 , -42.879581, -41.937173, -40.994764, -40.052356, -39.109948, -38.167539, -37.225131, -36.282723, -35.340314, -34.397906, -33.455497, -32.513089, -31.570681, -30.628272, -29.685864, -28.743455, -27.801047, -26.858639, -25.91623 , -24.973822, -24.031414, -23.089005, -22.146597, -21.204188, -20.26178 , -19.319372, -18.376963, -17.434555, -16.492147, -15.549738, -14.60733 , -13.664921, -12.722513, -11.780105, -10.837696, -9.895288, -8.95288 , -8.010471, -7.068063, -6.125654, -5.183246, -4.240838, -3.298429, -2.356021, -1.413613, -0.471204, 0.471204, 1.413613, 2.356021, 3.298429, 4.240838, 5.183246, 6.125654, 7.068063, 8.010471, 8.95288 , 9.895288, 10.837696, 11.780105, 12.722513, 13.664921, 14.60733 , 15.549738, 16.492147, 17.434555, 18.376963, 19.319372, 20.26178 , 21.204188, 22.146597, 23.089005, 24.031414, 24.973822, 25.91623 , 26.858639, 27.801047, 28.743455, 29.685864, 30.628272, 31.570681, 32.513089, 33.455497, 34.397906, 35.340314, 36.282723, 37.225131, 38.167539, 39.109948, 40.052356, 40.994764, 41.937173, 42.879581, 43.82199 , 44.764398, 45.706806, 46.649215, 47.591623, 48.534031, 49.47644 , 50.418848, 51.361257, 52.303665, 53.246073, 54.188482, 55.13089 , 56.073298, 57.015707, 57.958115, 58.900524, 59.842932, 60.78534 , 61.727749, 62.670157, 63.612565, 64.554974, 65.497382, 66.439791, 67.382199, 68.324607, 69.267016, 70.209424, 71.151832, 72.094241, 73.036649, 73.979058, 74.921466, 75.863874, 76.806283, 77.748691, 78.691099, 79.633508, 80.575916, 81.518325, 82.460733, 83.403141, 84.34555 , 85.287958, 86.230366, 87.172775, 88.115183, 89.057592, 90. ]) - lat_bnds(lat, nbnd)float32dask.array<chunksize=(192, 2), meta=np.ndarray>
- units :
- degrees_north
Array Chunk Bytes 1.54 kB 1.54 kB Shape (192, 2) (192, 2) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - lev(lev)float64-5.96e-06 -9.827e-06 ... -992.6
- axis :
- Z
- bounds :
- lev_bnds
- positive :
- up
- standard_name :
- alevel
- title :
- atmospheric model level
- type :
- double
- units :
- hPa
array([-5.960300e-06, -9.826900e-06, -1.620185e-05, -2.671225e-05, -4.404100e-05, -7.261275e-05, -1.197190e-04, -1.973800e-04, -3.254225e-04, -5.365325e-04, -8.846025e-04, -1.458457e-03, -2.404575e-03, -3.978250e-03, -6.556826e-03, -1.081383e-02, -1.789800e-02, -2.955775e-02, -4.873075e-02, -7.991075e-02, -1.282732e-01, -1.981200e-01, -2.920250e-01, -4.101675e-01, -5.534700e-01, -7.304800e-01, -9.559475e-01, -1.244795e+00, -1.612850e+00, -2.079325e+00, -2.667425e+00, -3.404875e+00, -4.324575e+00, -5.465400e+00, -6.872850e+00, -8.599725e+00, -1.070705e+01, -1.326475e+01, -1.635175e+01, -2.005675e+01, -2.447900e+01, -2.972800e+01, -3.592325e+01, -4.319375e+01, -5.167750e+01, -6.152050e+01, -7.375096e+01, -8.782123e+01, -1.033171e+02, -1.215472e+02, -1.429940e+02, -1.682251e+02, -1.979081e+02, -2.328286e+02, -2.739108e+02, -3.222419e+02, -3.791009e+02, -4.459926e+02, -5.246872e+02, -6.097787e+02, -6.913894e+02, -7.634045e+02, -8.208584e+02, -8.595348e+02, -8.870202e+02, -9.126445e+02, -9.361984e+02, -9.574855e+02, -9.763254e+02, -9.925561e+02]) - lev_bnds(lev, nbnd)float32dask.array<chunksize=(70, 2), meta=np.ndarray>
- formula :
- p = a*p0 + b*ps
- formula_terms :
- p0: p0 a: a_bnds b: b_bnds ps: ps
- standard_name :
- atmosphere_hybrid_sigma_pressure_coordinate
- units :
- hPa
Array Chunk Bytes 560 B 560 B Shape (70, 2) (70, 2) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - lon(lon)float640.0 1.25 2.5 ... 356.2 357.5 358.8
- axis :
- X
- bounds :
- lon_bnds
- standard_name :
- longitude
- title :
- Longitude
- type :
- double
- units :
- degrees_east
- valid_max :
- 360.0
- valid_min :
- 0.0
array([ 0. , 1.25, 2.5 , ..., 356.25, 357.5 , 358.75])
- lon_bnds(lon, nbnd)float32dask.array<chunksize=(288, 2), meta=np.ndarray>
- units :
- degrees_east
Array Chunk Bytes 2.30 kB 2.30 kB Shape (288, 2) (288, 2) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - time(time)object1850-01-15 12:00:00 ... 2014-12-...
- axis :
- T
- bounds :
- time_bnds
- standard_name :
- time
- title :
- time
- type :
- double
array([cftime.DatetimeNoLeap(1850, 1, 15, 12, 0, 0, 0), cftime.DatetimeNoLeap(1850, 2, 14, 0, 0, 0, 0), cftime.DatetimeNoLeap(1850, 3, 15, 12, 0, 0, 0), ..., cftime.DatetimeNoLeap(2014, 10, 15, 12, 0, 0, 0), cftime.DatetimeNoLeap(2014, 11, 15, 0, 0, 0, 0), cftime.DatetimeNoLeap(2014, 12, 15, 12, 0, 0, 0)], dtype=object) - time_bnds(time, nbnd)objectdask.array<chunksize=(1980, 2), meta=np.ndarray>
Array Chunk Bytes 31.68 kB 31.68 kB Shape (1980, 2) (1980, 2) Count 2 Tasks 1 Chunks Type object numpy.ndarray - member_id(member_id)<U8'r1i1p1f1'
array(['r1i1p1f1'], dtype='<U8')
- so2(member_id, time, lev, lat, lon)float32dask.array<chunksize=(1, 5, 70, 192, 288), meta=np.ndarray>
- cell_measures :
- area: areacella
- cell_methods :
- area: time: mean
- comment :
- Mole fraction is used in the construction mole_fraction_of_X_in_Y, where X is a material constituent of Y.
- description :
- Mole fraction is used in the construction mole_fraction_of_X_in_Y, where X is a material constituent of Y.
- frequency :
- mon
- id :
- so2
- long_name :
- SO2 Volume Mixing Ratio
- mipTable :
- AERmon
- out_name :
- so2
- prov :
- AERmon ((isd.003))
- realm :
- aerosol
- standard_name :
- mole_fraction_of_sulfur_dioxide_in_air
- time :
- time
- time_label :
- time-mean
- time_title :
- Temporal mean
- title :
- SO2 Volume Mixing Ratio
- type :
- real
- units :
- mol mol-1
- variable_id :
- so2
Array Chunk Bytes 30.66 GB 77.41 MB Shape (1, 1980, 70, 192, 288) (1, 5, 70, 192, 288) Count 793 Tasks 396 Chunks Type float32 numpy.ndarray
- Conventions :
- CF-1.7 CMIP-6.2
- activity_id :
- CMIP
- branch_method :
- standard
- branch_time_in_child :
- 674885.0
- branch_time_in_parent :
- 20075.0
- case_id :
- 4
- cesm_casename :
- b.e21.BWHIST.f09_g17.CMIP6-historical-WACCM.001
- contact :
- cesm_cmip6@ucar.edu
- creation_date :
- 2019-01-31T00:49:45Z
- data_specs_version :
- 01.00.29
- experiment :
- all-forcing simulation of the recent past
- experiment_id :
- historical
- external_variables :
- areacella
- forcing_index :
- 1
- frequency :
- mon
- further_info_url :
- https://furtherinfo.es-doc.org/CMIP6.NCAR.CESM2-WACCM.historical.none.r1i1p1f1
- grid :
- native 0.9x1.25 finite volume grid (192x288 latxlon)
- grid_label :
- gn
- initialization_index :
- 1
- institution :
- National Center for Atmospheric Research, Climate and Global Dynamics Laboratory, 1850 Table Mesa Drive, Boulder, CO 80305, USA
- institution_id :
- NCAR
- license :
- CMIP6 model data produced by <The National Center for Atmospheric Research> is licensed under a Creative Commons Attribution-[]ShareAlike 4.0 International License (https://creativecommons.org/licenses/). Consult https://pcmdi.llnl.gov/CMIP6/TermsOfUse for terms of use governing CMIP6 output, including citation requirements and proper acknowledgment. Further information about this data, including some limitations, can be found via the further_info_url (recorded as a global attribute in this file)[]. The data producers and data providers make no warranty, either express or implied, including, but not limited to, warranties of merchantability and fitness for a particular purpose. All liabilities arising from the supply of the information (including any liability arising in negligence) are excluded to the fullest extent permitted by law.
- mip_era :
- CMIP6
- model_doi_url :
- https://doi.org/10.5065/D67H1H0V
- nominal_resolution :
- 100 km
- parent_activity_id :
- CMIP
- parent_experiment_id :
- piControl
- parent_mip_era :
- CMIP6
- parent_source_id :
- CESM2-WACCM
- parent_time_units :
- days since 0001-01-01 00:00:00
- parent_variant_label :
- r1i1p1f1
- physics_index :
- 1
- product :
- model-output
- realization_index :
- 1
- realm :
- aerosol
- source :
- CESM2 (2017): atmosphere: CAM6 (0.9x1.25 finite volume grid; 288 x 192 longitude/latitude; 70 levels; top level 4.5e-6 mb); ocean: POP2 (320x384 longitude/latitude; 60 levels; top grid cell 0-10 m); sea_ice: CICE5.1 (same grid as ocean); land: CLM5 0.9x1.25 finite volume grid; 288 x 192 longitude/latitude; 70 levels; top level 4.5e-6 mb); aerosol: MAM4 (0.9x1.25 finite volume grid; 288 x 192 longitude/latitude; 70 levels; top level 4.5e-6 mb); atmosChem: WACCM (0.9x1.25 finite volume grid; 288 x 192 longitude/latitude; 70 levels; top level 4.5e-6 mb; landIce: CISM2.1; ocnBgchem: MARBL (320x384 longitude/latitude; 60 levels; top grid cell 0-10 m)
- source_id :
- CESM2-WACCM
- source_type :
- AOGCM BGC CHEM AER
- sub_experiment :
- none
- sub_experiment_id :
- none
- table_id :
- AERmon
- tracking_id :
- hdl:21.14100/c84179d5-78c4-4438-9849-d7a832efdc23
- variable_id :
- so2
- variant_info :
- CMIP6 CESM2 hindcast (1850-2014) with high-top atmosphere (WACCM6) with interactive chemistry (TSMLT1), interactive land (CLM5), coupled ocean (POP2) with biogeochemistry (MARBL), interactive sea ice (CICE5.1), and non-evolving land ice (CISM2.1)
- variant_label :
- r1i1p1f1
- status :
- 2019-11-05;created;by nhn2@columbia.edu
- intake_esm_varname :
- ['so2']
- intake_esm_dataset_key :
- CMIP.NCAR.CESM2-WACCM.historical.AERmon.gn
dset.so2
<xarray.DataArray 'so2' (member_id: 1, time: 1980, lev: 70, lat: 192, lon: 288)>
dask.array<broadcast_to, shape=(1, 1980, 70, 192, 288), dtype=float32, chunksize=(1, 5, 70, 192, 288), chunktype=numpy.ndarray>
Coordinates:
* lat (lat) float64 -90.0 -89.06 -88.12 -87.17 ... 88.12 89.06 90.0
* lev (lev) float64 -5.96e-06 -9.827e-06 -1.62e-05 ... -976.3 -992.6
* lon (lon) float64 0.0 1.25 2.5 3.75 5.0 ... 355.0 356.2 357.5 358.8
* time (time) object 1850-01-15 12:00:00 ... 2014-12-15 12:00:00
* member_id (member_id) <U8 'r1i1p1f1'
Attributes: (12/19)
cell_measures: area: areacella
cell_methods: area: time: mean
comment: Mole fraction is used in the construction mole_fraction_o...
description: Mole fraction is used in the construction mole_fraction_o...
frequency: mon
id: so2
... ...
time_label: time-mean
time_title: Temporal mean
title: SO2 Volume Mixing Ratio
type: real
units: mol mol-1
variable_id: so2xarray.DataArray
'so2'
- member_id: 1
- time: 1980
- lev: 70
- lat: 192
- lon: 288
- dask.array<chunksize=(1, 5, 70, 192, 288), meta=np.ndarray>
Array Chunk Bytes 30.66 GB 77.41 MB Shape (1, 1980, 70, 192, 288) (1, 5, 70, 192, 288) Count 793 Tasks 396 Chunks Type float32 numpy.ndarray - lat(lat)float64-90.0 -89.06 -88.12 ... 89.06 90.0
- axis :
- Y
- bounds :
- lat_bnds
- standard_name :
- latitude
- title :
- Latitude
- type :
- double
- units :
- degrees_north
- valid_max :
- 90.0
- valid_min :
- -90.0
array([-90. , -89.057592, -88.115183, -87.172775, -86.230366, -85.287958, -84.34555 , -83.403141, -82.460733, -81.518325, -80.575916, -79.633508, -78.691099, -77.748691, -76.806283, -75.863874, -74.921466, -73.979058, -73.036649, -72.094241, -71.151832, -70.209424, -69.267016, -68.324607, -67.382199, -66.439791, -65.497382, -64.554974, -63.612565, -62.670157, -61.727749, -60.78534 , -59.842932, -58.900524, -57.958115, -57.015707, -56.073298, -55.13089 , -54.188482, -53.246073, -52.303665, -51.361257, -50.418848, -49.47644 , -48.534031, -47.591623, -46.649215, -45.706806, -44.764398, -43.82199 , -42.879581, -41.937173, -40.994764, -40.052356, -39.109948, -38.167539, -37.225131, -36.282723, -35.340314, -34.397906, -33.455497, -32.513089, -31.570681, -30.628272, -29.685864, -28.743455, -27.801047, -26.858639, -25.91623 , -24.973822, -24.031414, -23.089005, -22.146597, -21.204188, -20.26178 , -19.319372, -18.376963, -17.434555, -16.492147, -15.549738, -14.60733 , -13.664921, -12.722513, -11.780105, -10.837696, -9.895288, -8.95288 , -8.010471, -7.068063, -6.125654, -5.183246, -4.240838, -3.298429, -2.356021, -1.413613, -0.471204, 0.471204, 1.413613, 2.356021, 3.298429, 4.240838, 5.183246, 6.125654, 7.068063, 8.010471, 8.95288 , 9.895288, 10.837696, 11.780105, 12.722513, 13.664921, 14.60733 , 15.549738, 16.492147, 17.434555, 18.376963, 19.319372, 20.26178 , 21.204188, 22.146597, 23.089005, 24.031414, 24.973822, 25.91623 , 26.858639, 27.801047, 28.743455, 29.685864, 30.628272, 31.570681, 32.513089, 33.455497, 34.397906, 35.340314, 36.282723, 37.225131, 38.167539, 39.109948, 40.052356, 40.994764, 41.937173, 42.879581, 43.82199 , 44.764398, 45.706806, 46.649215, 47.591623, 48.534031, 49.47644 , 50.418848, 51.361257, 52.303665, 53.246073, 54.188482, 55.13089 , 56.073298, 57.015707, 57.958115, 58.900524, 59.842932, 60.78534 , 61.727749, 62.670157, 63.612565, 64.554974, 65.497382, 66.439791, 67.382199, 68.324607, 69.267016, 70.209424, 71.151832, 72.094241, 73.036649, 73.979058, 74.921466, 75.863874, 76.806283, 77.748691, 78.691099, 79.633508, 80.575916, 81.518325, 82.460733, 83.403141, 84.34555 , 85.287958, 86.230366, 87.172775, 88.115183, 89.057592, 90. ]) - lev(lev)float64-5.96e-06 -9.827e-06 ... -992.6
- axis :
- Z
- bounds :
- lev_bnds
- positive :
- up
- standard_name :
- alevel
- title :
- atmospheric model level
- type :
- double
- units :
- hPa
array([-5.960300e-06, -9.826900e-06, -1.620185e-05, -2.671225e-05, -4.404100e-05, -7.261275e-05, -1.197190e-04, -1.973800e-04, -3.254225e-04, -5.365325e-04, -8.846025e-04, -1.458457e-03, -2.404575e-03, -3.978250e-03, -6.556826e-03, -1.081383e-02, -1.789800e-02, -2.955775e-02, -4.873075e-02, -7.991075e-02, -1.282732e-01, -1.981200e-01, -2.920250e-01, -4.101675e-01, -5.534700e-01, -7.304800e-01, -9.559475e-01, -1.244795e+00, -1.612850e+00, -2.079325e+00, -2.667425e+00, -3.404875e+00, -4.324575e+00, -5.465400e+00, -6.872850e+00, -8.599725e+00, -1.070705e+01, -1.326475e+01, -1.635175e+01, -2.005675e+01, -2.447900e+01, -2.972800e+01, -3.592325e+01, -4.319375e+01, -5.167750e+01, -6.152050e+01, -7.375096e+01, -8.782123e+01, -1.033171e+02, -1.215472e+02, -1.429940e+02, -1.682251e+02, -1.979081e+02, -2.328286e+02, -2.739108e+02, -3.222419e+02, -3.791009e+02, -4.459926e+02, -5.246872e+02, -6.097787e+02, -6.913894e+02, -7.634045e+02, -8.208584e+02, -8.595348e+02, -8.870202e+02, -9.126445e+02, -9.361984e+02, -9.574855e+02, -9.763254e+02, -9.925561e+02]) - lon(lon)float640.0 1.25 2.5 ... 356.2 357.5 358.8
- axis :
- X
- bounds :
- lon_bnds
- standard_name :
- longitude
- title :
- Longitude
- type :
- double
- units :
- degrees_east
- valid_max :
- 360.0
- valid_min :
- 0.0
array([ 0. , 1.25, 2.5 , ..., 356.25, 357.5 , 358.75])
- time(time)object1850-01-15 12:00:00 ... 2014-12-...
- axis :
- T
- bounds :
- time_bnds
- standard_name :
- time
- title :
- time
- type :
- double
array([cftime.DatetimeNoLeap(1850, 1, 15, 12, 0, 0, 0), cftime.DatetimeNoLeap(1850, 2, 14, 0, 0, 0, 0), cftime.DatetimeNoLeap(1850, 3, 15, 12, 0, 0, 0), ..., cftime.DatetimeNoLeap(2014, 10, 15, 12, 0, 0, 0), cftime.DatetimeNoLeap(2014, 11, 15, 0, 0, 0, 0), cftime.DatetimeNoLeap(2014, 12, 15, 12, 0, 0, 0)], dtype=object) - member_id(member_id)<U8'r1i1p1f1'
array(['r1i1p1f1'], dtype='<U8')
- cell_measures :
- area: areacella
- cell_methods :
- area: time: mean
- comment :
- Mole fraction is used in the construction mole_fraction_of_X_in_Y, where X is a material constituent of Y.
- description :
- Mole fraction is used in the construction mole_fraction_of_X_in_Y, where X is a material constituent of Y.
- frequency :
- mon
- id :
- so2
- long_name :
- SO2 Volume Mixing Ratio
- mipTable :
- AERmon
- out_name :
- so2
- prov :
- AERmon ((isd.003))
- realm :
- aerosol
- standard_name :
- mole_fraction_of_sulfur_dioxide_in_air
- time :
- time
- time_label :
- time-mean
- time_title :
- Temporal mean
- title :
- SO2 Volume Mixing Ratio
- type :
- real
- units :
- mol mol-1
- variable_id :
- so2
Compute Weighted average¶
# Compute weights based on the xarray you pass
weights = np.cos(np.deg2rad(dset.lat))
weights.name = "weights"
# Compute weighted mean
var_weighted = dset.sel(lev=-1000, method="nearest").weighted(weights)
weighted_mean = var_weighted.mean(("lon", "lat"))
Rolling mean¶
Choose rolling time of 100
%%time
weighted_mean.load()
CPU times: user 2min 51s, sys: 51.6 s, total: 3min 42s
Wall time: 7min 24s
<xarray.Dataset>
Dimensions: (member_id: 1, time: 1980)
Coordinates:
lev float64 -992.6
* time (time) object 1850-01-15 12:00:00 ... 2014-12-15 12:00:00
* member_id (member_id) <U8 'r1i1p1f1'
Data variables:
so2 (member_id, time) float64 5.779e-11 4.755e-11 ... 7.229e-10xarray.Dataset
- member_id: 1
- time: 1980
- lev()float64-992.6
- axis :
- Z
- bounds :
- lev_bnds
- positive :
- up
- standard_name :
- alevel
- title :
- atmospheric model level
- type :
- double
- units :
- hPa
array(-992.55609512)
- time(time)object1850-01-15 12:00:00 ... 2014-12-...
- axis :
- T
- bounds :
- time_bnds
- standard_name :
- time
- title :
- time
- type :
- double
array([cftime.DatetimeNoLeap(1850, 1, 15, 12, 0, 0, 0), cftime.DatetimeNoLeap(1850, 2, 14, 0, 0, 0, 0), cftime.DatetimeNoLeap(1850, 3, 15, 12, 0, 0, 0), ..., cftime.DatetimeNoLeap(2014, 10, 15, 12, 0, 0, 0), cftime.DatetimeNoLeap(2014, 11, 15, 0, 0, 0, 0), cftime.DatetimeNoLeap(2014, 12, 15, 12, 0, 0, 0)], dtype=object) - member_id(member_id)<U8'r1i1p1f1'
array(['r1i1p1f1'], dtype='<U8')
- so2(member_id, time)float645.779e-11 4.755e-11 ... 7.229e-10
array([[5.77863890e-11, 4.75515520e-11, 4.33568272e-11, ..., 4.82338678e-10, 6.06348895e-10, 7.22915999e-10]])
%%time
dpmean = weighted_mean.chunk(chunks={'time': 100}).rolling({'time':100},
min_periods=1,
center=True
).mean()
CPU times: user 65.7 ms, sys: 2.29 ms, total: 68 ms
Wall time: 66.3 ms
Visualize¶
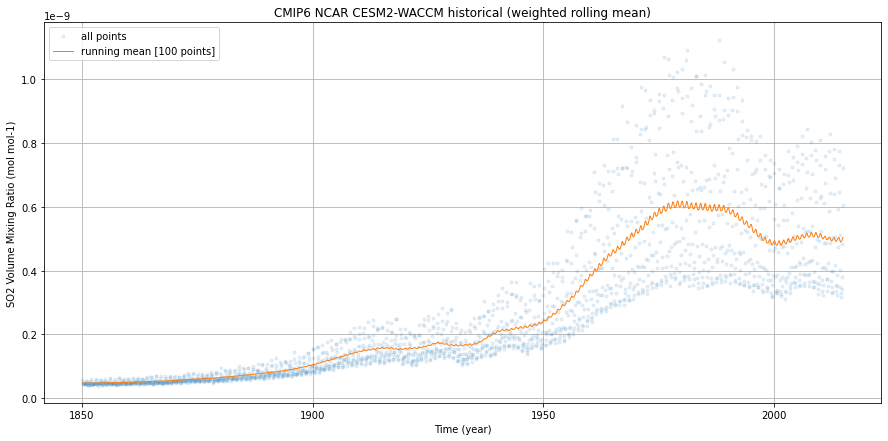
%%time
fig = plt.figure(1, figsize=[15,7])
ax = plt.subplot(1, 1, 1)
weighted_mean.so2.plot(ax=ax,
marker='.',
linewidth=0,
label = 'all points',
alpha=.1
)
dpmean.so2.plot(ax=ax,
marker='',
linewidth=1,
label = 'running mean [100 points]'
)
ax.legend()
ax.grid()
ax.set_ylabel(dset.so2.attrs['long_name'] + ' (' + dset.so2.attrs['units'] + ')')
ax.set_xlabel('Time (year)')
ax.set_title('CMIP6 NCAR CESM2-WACCM historical (weighted rolling mean)')
plt.savefig('CMIP_NCAR_CESM2-WACCM_historical_weighted_rolling_mean.png')
CPU times: user 570 ms, sys: 113 ms, total: 683 ms
Wall time: 682 ms
Save Results¶
To improve the reproducibility of our work (first for ourselves!), we need to keep track of all our work and ease its reuse. We will:
Save this Jupyter notebook
Save weighted mean
Save rolling mean
Save figure
We save intermediate results as netCDF files because they are small and ca be easily re-loaded (even if stored on cloud storage).
Save results locally¶
Useful for further analysis but can be lost if you close your JupyterLab or if there is any problem with your JupyterLab instance
weighted_mean.to_netcdf('CMIP_NCAR_CESM2-WACCM_historical_weighted_mean.nc')
dpmean.to_netcdf('CMIP_NCAR_CESM2-WACCM_historical_rolling_mean.nc')
Save your results on NIRD (Norwegian infrastructure for Research Data)¶
your credentials are in
$HOME/.aws/credentialscheck with your instructor to get the secret access key (replace XXX by the right key)
[default]
aws_access_key_id=forces2021-work
aws_secret_access_key=XXXXXXXXXXXX
aws_endpoint_url=https://forces2021.uiogeo-apps.sigma2.no/
It is important to save yoru results in a place that can last longer than a few days/weeks!
import s3fs
fsg = s3fs.S3FileSystem(anon=False,
client_kwargs={
'endpoint_url': 'https://forces2021.uiogeo-apps.sigma2.no/'
})
Set “remote” path (update annefou by your username) and save weighted mean as netCDF file¶
s3_path = "s3://work/annefou/CMIP_NCAR_CESM2-WACCM_historical_weighted_mean.nc"
print(s3_path)
s3://work/annefou/CMIP_NCAR_CESM2-WACCM_historical_weighted_mean.nc
with fsg.open(s3_path, 'wb') as f:
f.write(weighted_mean.to_netcdf(None))
Save rolling mean to remote location (update annefou with your username)¶
s3_path = "s3://work/annefou/CMIP_NCAR_CESM2-WACCM_historical_rolling_mean.nc"
print(s3_path)
s3://work/annefou/CMIP_NCAR_CESM2-WACCM_historical_rolling_mean.nc
with fsg.open(s3_path, 'wb') as f:
f.write(dpmean.to_netcdf(None))
Upload existing png file to remote s3 location¶
s3_path = "s3://work/annefou/CMIP_NCAR_CESM2-WACCM_historical_weighted_rolling_mean.png"
print(s3_path)
s3://work/annefou/CMIP_NCAR_CESM2-WACCM_historical_weighted_rolling_mean.png
fsg.put('CMIP_NCAR_CESM2-WACCM_historical_weighted_rolling_mean.png', s3_path)
