Multidimensional Coordinates example using CMIP6 Pangeo ocean model data
Contents
Multidimensional Coordinates example using CMIP6 Pangeo ocean model data¶
Import python packages¶
import xarray as xr
xr.set_options(display_style='html')
import intake
import cftime
import matplotlib.pyplot as plt
import matplotlib.path as mpath
import cartopy.crs as ccrs
import numpy as np
%matplotlib inline
Open CMIP6 online catalog¶
cat_url = "https://storage.googleapis.com/cmip6/pangeo-cmip6.json"
col = intake.open_esm_datastore(cat_url)
col
pangeo-cmip6 catalog with 7715 dataset(s) from 520279 asset(s):
| unique | |
|---|---|
| activity_id | 18 |
| institution_id | 36 |
| source_id | 88 |
| experiment_id | 170 |
| member_id | 657 |
| table_id | 37 |
| variable_id | 709 |
| grid_label | 10 |
| zstore | 520279 |
| dcpp_init_year | 60 |
| version | 723 |
Search corresponding data¶
cat = col.search(source_id=['MPI-ESM1-2-HR'], experiment_id=['historical'], table_id=['Omon'], variable_id=['chl'], member_id=['r9i1p1f1'])
cat.df
| activity_id | institution_id | source_id | experiment_id | member_id | table_id | variable_id | grid_label | zstore | dcpp_init_year | version | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | CMIP | MPI-M | MPI-ESM1-2-HR | historical | r9i1p1f1 | Omon | chl | gn | gs://cmip6/CMIP6/CMIP/MPI-M/MPI-ESM1-2-HR/hist... | NaN | 20190710 |
Create dictionary from the list of datasets we found¶
This step may take several minutes so be patient!
dset_dict = cat.to_dataset_dict(zarr_kwargs={'use_cftime':True})
--> The keys in the returned dictionary of datasets are constructed as follows:
'activity_id.institution_id.source_id.experiment_id.table_id.grid_label'
100.00% [1/1 00:00<00:00]
list(dset_dict.keys())
['CMIP.MPI-M.MPI-ESM1-2-HR.historical.Omon.gn']
Open dataset¶
Use
xarraypython package to analyze netCDF datasetopen_datasetallows to get all the metadata without loading data into memory.with
xarray, we only load into memory what is needed.
dset = dset_dict['CMIP.MPI-M.MPI-ESM1-2-HR.historical.Omon.gn']
dset
<xarray.Dataset>
Dimensions: (bnds: 2, i: 802, j: 404, lev: 40, member_id: 1, time: 1980, vertices: 4)
Coordinates:
* i (i) int32 0 1 2 3 4 5 6 ... 795 796 797 798 799 800 801
* j (j) int32 0 1 2 3 4 5 6 ... 397 398 399 400 401 402 403
latitude (j, i) float64 dask.array<chunksize=(404, 802), meta=np.ndarray>
* lev (lev) float64 6.0 17.0 27.0 ... 5.17e+03 5.72e+03
lev_bnds (lev, bnds) float64 dask.array<chunksize=(40, 2), meta=np.ndarray>
longitude (j, i) float64 dask.array<chunksize=(404, 802), meta=np.ndarray>
* time (time) object 1850-01-16 12:00:00 ... 2014-12-16 12:0...
time_bnds (time, bnds) object dask.array<chunksize=(1980, 2), meta=np.ndarray>
* member_id (member_id) <U8 'r9i1p1f1'
Dimensions without coordinates: bnds, vertices
Data variables:
chl (member_id, time, lev, j, i) float32 dask.array<chunksize=(1, 2, 40, 404, 802), meta=np.ndarray>
vertices_latitude (j, i, vertices) float64 dask.array<chunksize=(404, 802, 4), meta=np.ndarray>
vertices_longitude (j, i, vertices) float64 dask.array<chunksize=(404, 802, 4), meta=np.ndarray>
Attributes: (12/50)
Conventions: CF-1.7 CMIP-6.2
activity_id: CMIP
branch_method: standard
branch_time_in_child: 0.0
branch_time_in_parent: 146096.0
cmor_version: 3.5.0
... ...
title: MPI-ESM1-2-HR output prepared for CMIP6
tracking_id: hdl:21.14100/4e922d88-7711-4df6-9bdd-b849acd45a7...
variable_id: chl
variant_label: r9i1p1f1
intake_esm_varname: ['chl']
intake_esm_dataset_key: CMIP.MPI-M.MPI-ESM1-2-HR.historical.Omon.gnxarray.Dataset
- bnds: 2
- i: 802
- j: 404
- lev: 40
- member_id: 1
- time: 1980
- vertices: 4
- i(i)int320 1 2 3 4 5 ... 797 798 799 800 801
- long_name :
- cell index along first dimension
- units :
- 1
array([ 0, 1, 2, ..., 799, 800, 801], dtype=int32)
- j(j)int320 1 2 3 4 5 ... 399 400 401 402 403
- long_name :
- cell index along second dimension
- units :
- 1
array([ 0, 1, 2, ..., 401, 402, 403], dtype=int32)
- latitude(j, i)float64dask.array<chunksize=(404, 802), meta=np.ndarray>
- bounds :
- vertices_latitude
- long_name :
- latitude
- standard_name :
- latitude
- units :
- degrees_north
Array Chunk Bytes 2.59 MB 2.59 MB Shape (404, 802) (404, 802) Count 2 Tasks 1 Chunks Type float64 numpy.ndarray - lev(lev)float646.0 17.0 27.0 ... 5.17e+03 5.72e+03
- axis :
- Z
- bounds :
- lev_bnds
- long_name :
- ocean depth coordinate
- positive :
- down
- standard_name :
- depth
- units :
- m
array([ 6. , 17. , 27. , 37. , 47. , 57. , 68.5, 82.5, 100. , 122.5, 150. , 182.5, 220. , 262.5, 310. , 362.5, 420. , 485. , 560. , 645. , 740. , 845. , 960. , 1085. , 1220. , 1365. , 1525. , 1700. , 1885. , 2080. , 2290. , 2525. , 2785. , 3070. , 3395. , 3770. , 4195. , 4670. , 5170. , 5720. ]) - lev_bnds(lev, bnds)float64dask.array<chunksize=(40, 2), meta=np.ndarray>
Array Chunk Bytes 640 B 640 B Shape (40, 2) (40, 2) Count 2 Tasks 1 Chunks Type float64 numpy.ndarray - longitude(j, i)float64dask.array<chunksize=(404, 802), meta=np.ndarray>
- bounds :
- vertices_longitude
- long_name :
- longitude
- standard_name :
- longitude
- units :
- degrees_east
Array Chunk Bytes 2.59 MB 2.59 MB Shape (404, 802) (404, 802) Count 2 Tasks 1 Chunks Type float64 numpy.ndarray - time(time)object1850-01-16 12:00:00 ... 2014-12-...
- axis :
- T
- bounds :
- time_bnds
- long_name :
- time
- standard_name :
- time
array([cftime.DatetimeProlepticGregorian(1850, 1, 16, 12, 0, 0, 0), cftime.DatetimeProlepticGregorian(1850, 2, 15, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(1850, 3, 16, 12, 0, 0, 0), ..., cftime.DatetimeProlepticGregorian(2014, 10, 16, 12, 0, 0, 0), cftime.DatetimeProlepticGregorian(2014, 11, 16, 0, 0, 0, 0), cftime.DatetimeProlepticGregorian(2014, 12, 16, 12, 0, 0, 0)], dtype=object) - time_bnds(time, bnds)objectdask.array<chunksize=(1980, 2), meta=np.ndarray>
Array Chunk Bytes 31.68 kB 31.68 kB Shape (1980, 2) (1980, 2) Count 2 Tasks 1 Chunks Type object numpy.ndarray - member_id(member_id)<U8'r9i1p1f1'
array(['r9i1p1f1'], dtype='<U8')
- chl(member_id, time, lev, j, i)float32dask.array<chunksize=(1, 2, 40, 404, 802), meta=np.ndarray>
- cell_measures :
- area: areacello volume: volcello
- cell_methods :
- area: mean where sea time: mean
- comment :
- Sum of chlorophyll from all phytoplankton group concentrations. In most models this is equal to chldiat+chlmisc, that is the sum of Diatom Chlorophyll Mass Concentration and Other Phytoplankton Chlorophyll Mass Concentration
- history :
- 2019-08-30T07:10:19Z altered by CMOR: replaced missing value flag (-9e+33) and corresponding data with standard missing value (1e+20).
- long_name :
- Mass Concentration of Total Phytoplankton Expressed as Chlorophyll in Sea Water
- original_name :
- chl
- standard_name :
- mass_concentration_of_phytoplankton_expressed_as_chlorophyll_in_sea_water
- units :
- kg m-3
Array Chunk Bytes 102.65 GB 103.68 MB Shape (1, 1980, 40, 404, 802) (1, 2, 40, 404, 802) Count 1981 Tasks 990 Chunks Type float32 numpy.ndarray - vertices_latitude(j, i, vertices)float64dask.array<chunksize=(404, 802, 4), meta=np.ndarray>
- units :
- degrees_north
Array Chunk Bytes 10.37 MB 10.37 MB Shape (404, 802, 4) (404, 802, 4) Count 2 Tasks 1 Chunks Type float64 numpy.ndarray - vertices_longitude(j, i, vertices)float64dask.array<chunksize=(404, 802, 4), meta=np.ndarray>
- units :
- degrees_east
Array Chunk Bytes 10.37 MB 10.37 MB Shape (404, 802, 4) (404, 802, 4) Count 2 Tasks 1 Chunks Type float64 numpy.ndarray
- Conventions :
- CF-1.7 CMIP-6.2
- activity_id :
- CMIP
- branch_method :
- standard
- branch_time_in_child :
- 0.0
- branch_time_in_parent :
- 146096.0
- cmor_version :
- 3.5.0
- contact :
- cmip6-mpi-esm@dkrz.de
- creation_date :
- 2019-08-30T07:10:20Z
- data_specs_version :
- 01.00.30
- experiment :
- all-forcing simulation of the recent past
- experiment_id :
- historical
- external_variables :
- areacello volcello
- forcing_index :
- 1
- frequency :
- mon
- further_info_url :
- https://furtherinfo.es-doc.org/CMIP6.MPI-M.MPI-ESM1-2-HR.historical.none.r9i1p1f1
- grid :
- gn
- grid_label :
- gn
- history :
- 2019-08-30T07:10:20Z ; CMOR rewrote data to be consistent with CMIP6, CF-1.7 CMIP-6.2 and CF standards.
- initialization_index :
- 1
- institution :
- Max Planck Institute for Meteorology, Hamburg 20146, Germany
- institution_id :
- MPI-M
- license :
- CMIP6 model data produced by MPI-M is licensed under a Creative Commons Attribution ShareAlike 4.0 International License (https://creativecommons.org/licenses). Consult https://pcmdi.llnl.gov/CMIP6/TermsOfUse for terms of use governing CMIP6 output, including citation requirements and proper acknowledgment. Further information about this data, including some limitations, can be found via the further_info_url (recorded as a global attribute in this file) and. The data producers and data providers make no warranty, either express or implied, including, but not limited to, warranties of merchantability and fitness for a particular purpose. All liabilities arising from the supply of the information (including any liability arising in negligence) are excluded to the fullest extent permitted by law.
- mip_era :
- CMIP6
- nominal_resolution :
- 50 km
- parent_activity_id :
- CMIP
- parent_experiment_id :
- piControl
- parent_mip_era :
- CMIP6
- parent_source_id :
- MPI-ESM1-2-HR
- parent_time_units :
- days since 1850-1-1 00:00:00
- parent_variant_label :
- r9i1p1f1
- physics_index :
- 1
- product :
- model-output
- project_id :
- CMIP6
- realization_index :
- 9
- realm :
- ocnBgchem
- references :
- MPI-ESM: Mauritsen, T. et al. (2019), Developments in the MPI‐M Earth System Model version 1.2 (MPI‐ESM1.2) and Its Response to Increasing CO2, J. Adv. Model. Earth Syst.,11, 998-1038, doi:10.1029/2018MS001400, Mueller, W.A. et al. (2018): A high‐resolution version of the Max Planck Institute Earth System Model MPI‐ESM1.2‐HR. J. Adv. Model. EarthSyst.,10,1383–1413, doi:10.1029/2017MS001217
- source :
- MPI-ESM1.2-HR (2017): aerosol: none, prescribed MACv2-SP atmos: ECHAM6.3 (spectral T127; 384 x 192 longitude/latitude; 95 levels; top level 0.01 hPa) atmosChem: none land: JSBACH3.20 landIce: none/prescribed ocean: MPIOM1.63 (tripolar TP04, approximately 0.4deg; 802 x 404 longitude/latitude; 40 levels; top grid cell 0-12 m) ocnBgchem: HAMOCC6 seaIce: unnamed (thermodynamic (Semtner zero-layer) dynamic (Hibler 79) sea ice model)
- source_id :
- MPI-ESM1-2-HR
- source_type :
- AOGCM
- status :
- 2020-04-18;created; by gcs.cmip6.ldeo@gmail.com
- sub_experiment :
- none
- sub_experiment_id :
- none
- table_id :
- Omon
- table_info :
- Creation Date:(09 May 2019) MD5:e6ef8ececc8f338646ebfb3aeed36bfc
- title :
- MPI-ESM1-2-HR output prepared for CMIP6
- tracking_id :
- hdl:21.14100/4e922d88-7711-4df6-9bdd-b849acd45a77 hdl:21.14100/c80d8b77-125c-4d40-84d8-5d96d26c8871 hdl:21.14100/256ed3bd-05a0-4e98-bbbb-d45570361013 hdl:21.14100/985630f9-9922-41ed-8cfe-2e5dbe49a5ee hdl:21.14100/07ae37ed-16a0-4412-96b0-d2625894c68f hdl:21.14100/9c36da05-c73c-4259-baf5-76d3877b5cfc hdl:21.14100/20715c6d-d9f8-483e-922c-05374cac92ae hdl:21.14100/33ca1735-dfbb-4d5c-8457-164d34583d2d hdl:21.14100/39619a6e-af06-4fe8-99ec-bb442024599a hdl:21.14100/cbad2b59-fba5-49b8-aa09-106afb600812 hdl:21.14100/162c662d-839d-430a-8261-0ac73f4643d0 hdl:21.14100/1c87d835-d9c9-4458-bd52-a972498fcfb8 hdl:21.14100/ea60206c-889c-4f00-b295-daf4a4f02d96 hdl:21.14100/589eea2f-deef-439e-a2e1-afa4a506fcc7 hdl:21.14100/eba1472c-3029-4be3-acdf-288c38c86336 hdl:21.14100/48b01132-09f5-4f45-b05b-e10a41eb29d8 hdl:21.14100/c63ce8ba-967f-4827-afa9-3339d22d9d44 hdl:21.14100/dd2dbac5-8c70-4902-8f52-b8668684da80 hdl:21.14100/434b1bcc-de97-437e-b1f8-b468c054e819 hdl:21.14100/d1fc1095-0263-4231-884b-67b6b3aa3248 hdl:21.14100/a1ce2b6c-0787-4cf2-adc3-9c53f5577d05 hdl:21.14100/60bff6a6-ffde-4604-a53a-89442b8f0fc7 hdl:21.14100/b57d4410-a2c2-41a5-bdc8-ceeb9ae553c3 hdl:21.14100/b96a67c6-c1ce-4603-b528-665880ca4288 hdl:21.14100/63f99a97-4708-4063-ae9d-13dc81a6b8d4 hdl:21.14100/4a3f98bd-a2ff-4fbb-99a8-9ad632810b75 hdl:21.14100/b9c209c3-29e3-47b2-889c-104de3f08297 hdl:21.14100/33b509f0-f0c6-48b3-a342-62b727a50bae hdl:21.14100/4c9dbc22-a1fc-4a8f-aa74-6cf062b232c7 hdl:21.14100/fb0ac57b-b06b-4d0c-b322-4b90434a4b6a hdl:21.14100/00ba5a3f-281e-4bea-a673-4b65e9d60c93 hdl:21.14100/7e50067b-045e-4dda-b221-b68f57c831c0 hdl:21.14100/ca3ddf73-a346-488f-a862-1f1db4a9448b
- variable_id :
- chl
- variant_label :
- r9i1p1f1
- intake_esm_varname :
- ['chl']
- intake_esm_dataset_key :
- CMIP.MPI-M.MPI-ESM1-2-HR.historical.Omon.gn
Get metadata corresponding to Chl¶
print(dset['chl'])
<xarray.DataArray 'chl' (member_id: 1, time: 1980, lev: 40, j: 404, i: 802)>
dask.array<broadcast_to, shape=(1, 1980, 40, 404, 802), dtype=float32, chunksize=(1, 2, 40, 404, 802), chunktype=numpy.ndarray>
Coordinates:
* i (i) int32 0 1 2 3 4 5 6 7 8 ... 794 795 796 797 798 799 800 801
* j (j) int32 0 1 2 3 4 5 6 7 8 ... 396 397 398 399 400 401 402 403
latitude (j, i) float64 dask.array<chunksize=(404, 802), meta=np.ndarray>
* lev (lev) float64 6.0 17.0 27.0 37.0 ... 4.67e+03 5.17e+03 5.72e+03
longitude (j, i) float64 dask.array<chunksize=(404, 802), meta=np.ndarray>
* time (time) object 1850-01-16 12:00:00 ... 2014-12-16 12:00:00
* member_id (member_id) <U8 'r9i1p1f1'
Attributes:
cell_measures: area: areacello volume: volcello
cell_methods: area: mean where sea time: mean
comment: Sum of chlorophyll from all phytoplankton group concentra...
history: 2019-08-30T07:10:19Z altered by CMOR: replaced missing va...
long_name: Mass Concentration of Total Phytoplankton Expressed as Ch...
original_name: chl
standard_name: mass_concentration_of_phytoplankton_expressed_as_chloroph...
units: kg m-3
dset.time.values
array([cftime.DatetimeProlepticGregorian(1850, 1, 16, 12, 0, 0, 0),
cftime.DatetimeProlepticGregorian(1850, 2, 15, 0, 0, 0, 0),
cftime.DatetimeProlepticGregorian(1850, 3, 16, 12, 0, 0, 0), ...,
cftime.DatetimeProlepticGregorian(2014, 10, 16, 12, 0, 0, 0),
cftime.DatetimeProlepticGregorian(2014, 11, 16, 0, 0, 0, 0),
cftime.DatetimeProlepticGregorian(2014, 12, 16, 12, 0, 0, 0)],
dtype=object)
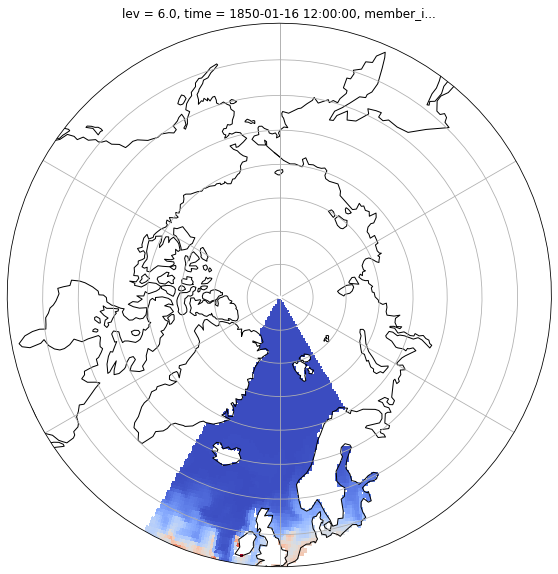
Visualization¶
shift longitude from 0,360 to -180,180¶
dset.coords['longitude'] = (dset['longitude'] + 180) % 360 - 180
dset['chl'].latitude.values.min(), dset['chl'].latitude.values.max()
(-78.71779338013427, 89.76311914726304)
dset['chl'].longitude.values.min(), dset['chl'].longitude.values.max()
(-179.99408443971072, 179.99893486577713)
Select geographical area, time and level¶
dset_selection = dset.isel(time=0, lev=0).where( (dset.latitude > 50) & (dset.longitude > -30) & (dset.longitude < 30) ).squeeze()
def polarCentral_set_latlim(lat_lims, ax):
ax.set_extent([-180, 180, lat_lims[0], lat_lims[1]], ccrs.PlateCarree())
# Compute a circle in axes coordinates, which we can use as a boundary
# for the map. We can pan/zoom as much as we like - the boundary will be
# permanently circular.
theta = np.linspace(0, 2*np.pi, 100)
center, radius = [0.5, 0.5], 0.5
verts = np.vstack([np.sin(theta), np.cos(theta)]).T
circle = mpath.Path(verts * radius + center)
ax.set_boundary(circle, transform=ax.transAxes)
fig = plt.figure(1, figsize=[10,10])
# Fix extent
minval = 0
maxval = 100.
ax = plt.subplot(1, 1, 1, projection=ccrs.NorthPolarStereo())
ax.coastlines()
ax.gridlines()
polarCentral_set_latlim([50,90], ax)
dset_selection.chl.plot.pcolormesh(
ax=ax, transform=ccrs.PlateCarree(), x="longitude", y="latitude", add_colorbar=False, cmap='coolwarm')
/opt/conda/lib/python3.8/site-packages/cartopy/mpl/geoaxes.py:1597: UserWarning: The input coordinates to pcolormesh are interpreted as cell centers, but are not monotonically increasing or decreasing. This may lead to incorrectly calculated cell edges, in which case, please supply explicit cell edges to pcolormesh.
X, Y, C, shading = self._pcolorargs('pcolormesh', *args,
<matplotlib.collections.QuadMesh at 0x7f109bda9b80>