Using ADAM-API to access MODIS Aqua CHL
Contents
Using ADAM-API to access MODIS Aqua CHL¶
you need to get an account to https://reliance.adamplatform.eu/ (use ORCID to authenticate) and key your ADAM API key
make sure you save your ADAM API key in a file
$HOME/adam-key
!pip install adamapi
WARNING: The directory '/home/jovyan/.cache/pip' or its parent directory is not owned or is not writable by the current user. The cache has been disabled. Check the permissions and owner of that directory. If executing pip with sudo, you may want sudo's -H flag.
Collecting adamapi
Downloading adamapi-2.0.11-py3-none-any.whl (14 kB)
Requirement already satisfied: requests>=2.22.0 in /srv/conda/envs/notebook/lib/python3.9/site-packages (from adamapi) (2.26.0)
Requirement already satisfied: imageio in /srv/conda/envs/notebook/lib/python3.9/site-packages (from adamapi) (2.9.0)
Requirement already satisfied: certifi>=2017.4.17 in /srv/conda/envs/notebook/lib/python3.9/site-packages (from requests>=2.22.0->adamapi) (2021.10.8)
Requirement already satisfied: idna<4,>=2.5 in /srv/conda/envs/notebook/lib/python3.9/site-packages (from requests>=2.22.0->adamapi) (3.1)
Requirement already satisfied: urllib3<1.27,>=1.21.1 in /srv/conda/envs/notebook/lib/python3.9/site-packages (from requests>=2.22.0->adamapi) (1.26.7)
Requirement already satisfied: charset-normalizer~=2.0.0 in /srv/conda/envs/notebook/lib/python3.9/site-packages (from requests>=2.22.0->adamapi) (2.0.0)
Requirement already satisfied: numpy in /srv/conda/envs/notebook/lib/python3.9/site-packages (from imageio->adamapi) (1.20.0)
Requirement already satisfied: pillow in /srv/conda/envs/notebook/lib/python3.9/site-packages (from imageio->adamapi) (8.4.0)
Installing collected packages: adamapi
Successfully installed adamapi-2.0.11
!pip install cmocean
WARNING: The directory '/home/jovyan/.cache/pip' or its parent directory is not owned or is not writable by the current user. The cache has been disabled. Check the permissions and owner of that directory. If executing pip with sudo, you may want sudo's -H flag.
Collecting cmocean
Downloading cmocean-2.0-py3-none-any.whl (223 kB)
|████████████████████████████████| 223 kB 5.5 MB/s eta 0:00:01
?25hInstalling collected packages: cmocean
Successfully installed cmocean-2.0
import os
import glob
import pathlib
import zipfile
import adamapi as adam
import xarray as xr
from datetime import datetime
import matplotlib
import matplotlib.pyplot as plt
import cartopy.crs as ccrs
import cmocean
Authenticate to ADAM platform with ADAM API key¶
adam_key = open(os.path.join(os.environ['HOME'],"adam-key")).read().rstrip()
a = adam.Auth()
a.setKey(adam_key)
a.setAdamCore('https://reliance.adamplatform.eu')
a.authorize()
{'expires_at': '2021-12-08T20:07:46.752Z',
'access_token': 'a34db20c230447059c2cf7586c3e5d63',
'refresh_token': '17aba7b3192c4f27b47837e93449e4c7',
'expires_in': 3600}
Discover MOD_Aqua datasets¶
This step is useful to get the dataset identifier (unique for a given datacube)
def discoverDasasets(a, search_name):
datasets = adam.Datasets(a)
catalogue = datasets.getDatasets()
#Extracting the size of the catalogue
total = catalogue['properties']['totalResults']
items = catalogue['properties']['itemsPerPage']
pages = total//items
print('----------------------------------------------------------------------')
print('\033[1m' + 'List of available datasets:')
print ('\033[0m')
#Extracting the list of datasets across the whole catalogue
for i in range(0,pages):
page = datasets.getDatasets(page = i)
for element in page['content']:
if search_name in element['title'] :
print(element['title'] + "\033[1m" + " --> datasetId "+ "\033[0m" + "= " + element['datasetId'])
return datasets
datasets = discoverDasasets(a, 'MOD_Aqua')
----------------------------------------------------------------------
List of available datasets:
Get metadata from Modis Aqua Chlorophylle Concentration¶
datasetID = '69618:MODh20chlMO_4km'
print('\033[1;34m' + 'Metadata of ' + datasetID + ':')
print ('\033[0;0m')
paged = datasets.getDatasets(datasetID)
for i in paged.items():
print("\033[1m" + str(i[0]) + "\033[0m" + ': ' + str(i[1]))
Metadata of 69618:MODh20chlMO_4km:
datasetId: 69618:MODh20chlMO_4km
creationDate: 2018-12-01T00:10:01Z
dataType: Float32
epsg: 4326
keywords: ['/processing_control/input_parameters/NC_GLOBAL#alt_thresh_lat=999', '/processing_control/input_parameters/NC_GLOBAL#apply_pal=1', '/processing_control/input_parameters/NC_GLOBAL#central_meridian=-999', '/processing_control/input_parameters/NC_GLOBAL#deflate=4', '/processing_control/input_parameters/NC_GLOBAL#east=180.000', '/processing_control/input_parameters/NC_GLOBAL#fudge=1.0', '/processing_control/input_parameters/NC_GLOBAL#full_latlon=yes', '/processing_control/input_parameters/NC_GLOBAL#interp=area', '/processing_control/input_parameters/NC_GLOBAL#land=$OCDATAROOT/common/landmask_GMT15ARC.nc', '/processing_control/input_parameters/NC_GLOBAL#mask_land=no', '/processing_control/input_parameters/NC_GLOBAL#north=90.000', '/processing_control/input_parameters/NC_GLOBAL#num_cache=500', '/processing_control/input_parameters/NC_GLOBAL#oformat=2', '/processing_control/input_parameters/NC_GLOBAL#oformat2=png', '/processing_control/input_parameters/NC_GLOBAL#product=chlor_a', '/processing_control/input_parameters/NC_GLOBAL#product_rgb=rhos_645,rhos_555,rhos_469', '/processing_control/input_parameters/NC_GLOBAL#projection=smi', '/processing_control/input_parameters/NC_GLOBAL#quiet=false', '/processing_control/input_parameters/NC_GLOBAL#resolution=4km', '/processing_control/input_parameters/NC_GLOBAL#south=-90.000', '/processing_control/input_parameters/NC_GLOBAL#suite=CHL', '/processing_control/input_parameters/NC_GLOBAL#threshold=0', '/processing_control/input_parameters/NC_GLOBAL#trimNSEW=yes', '/processing_control/input_parameters/NC_GLOBAL#use_quality=yes', '/processing_control/input_parameters/NC_GLOBAL#use_rgb=no', '/processing_control/input_parameters/NC_GLOBAL#west=-180.000', '/processing_control/NC_GLOBAL#l2_flag_names=ATMFAIL,LAND,HILT,HISATZEN,STRAYLIGHT,CLDICE,COCCOLITH,LOWLW,CHLWARN,CHLFAIL,NAVWARN,MAXAERITER,ATMWARN,HISOLZEN,NAVFAIL,FILTER,HIGLINT', '/processing_control/NC_GLOBAL#software_name=l3mapgen', '/processing_control/NC_GLOBAL#software_version=2.1.0-V2018.2.1', 'chlor_a#display_max=20', 'chlor_a#display_min=0.0099999998', 'chlor_a#display_scale=log', 'chlor_a#long_name=Chlorophyll Concentration, OCI Algorithm', 'chlor_a#reference=Hu, C., Lee Z., and Franz, B.A. (2012). Chlorophyll-a algorithms for oligotrophic oceans: A novel approach based on three-band reflectance difference, J. Geophys. Res., 117, C01011, doi:10.1029/2011JC007395.', 'chlor_a#standard_name=mass_concentration_of_chlorophyll_in_sea_water', 'chlor_a#units=mg m^-3', 'chlor_a#valid_max=100', 'chlor_a#valid_min=0.001', 'chlor_a#_FillValue=-32767', 'lat#long_name=Latitude', 'lat#standard_name=latitude', 'lat#units=degrees_north', 'lat#valid_max=90', 'lat#valid_min=-90', 'lat#_FillValue=-999', 'lon#long_name=Longitude', 'lon#standard_name=longitude', 'lon#units=degrees_east', 'lon#valid_max=180', 'lon#valid_min=-180', 'lon#_FillValue=-999', 'NC_GLOBAL#cdm_data_type=grid', 'NC_GLOBAL#Conventions=CF-1.6 ACDD-1.3', 'NC_GLOBAL#creator_email=data@oceancolor.gsfc.nasa.gov', 'NC_GLOBAL#creator_name=NASA/GSFC/OBPG', 'NC_GLOBAL#creator_url=http://oceandata.sci.gsfc.nasa.gov', 'NC_GLOBAL#data_bins=16635770', 'NC_GLOBAL#data_maximum=99.822014', 'NC_GLOBAL#data_minimum=0.0061450284', 'NC_GLOBAL#easternmost_longitude=180', 'NC_GLOBAL#end_orbit_number=88625', 'NC_GLOBAL#geospatial_lat_max=90', 'NC_GLOBAL#geospatial_lat_min=-90', 'NC_GLOBAL#geospatial_lat_resolution=4.6383123', 'NC_GLOBAL#geospatial_lat_units=degrees_north', 'NC_GLOBAL#geospatial_lon_max=180', 'NC_GLOBAL#geospatial_lon_min=-180', 'NC_GLOBAL#geospatial_lon_resolution=4.6383123', 'NC_GLOBAL#geospatial_lon_units=degrees_east', 'NC_GLOBAL#identifier_product_doi_authority=http://dx.doi.org', 'NC_GLOBAL#institution=NASA Goddard Space Flight Center, Ocean Ecology Laboratory, Ocean Biology Processing Group', 'NC_GLOBAL#instrument=MODIS', 'NC_GLOBAL#keywords=Earth Science > Oceans > Ocean Chemistry > Pigments > Chlorophyll; Earth Science > Oceans > Ocean Chemistry > Chlorophyllr', 'NC_GLOBAL#keywords_vocabulary=NASA Global Change Master Directory (GCMD) Science Keywords', 'NC_GLOBAL#l2_flag_names=ATMFAIL,LAND,HILT,HISATZEN,STRAYLIGHT,CLDICE,COCCOLITH,LOWLW,CHLWARN,CHLFAIL,NAVWARN,MAXAERITER,ATMWARN,HISOLZEN,NAVFAIL,FILTER,HIGLINT', 'NC_GLOBAL#latitude_step=0.041666668', 'NC_GLOBAL#latitude_units=degrees_north', 'NC_GLOBAL#license=http://science.nasa.gov/earth-science/earth-science-data/data-information-policy/', 'NC_GLOBAL#longitude_step=0.041666668', 'NC_GLOBAL#longitude_units=degrees_east', 'NC_GLOBAL#map_projection=Equidistant Cylindrical', 'NC_GLOBAL#measure=Mean', 'NC_GLOBAL#naming_authority=gov.nasa.gsfc.sci.oceandata', 'NC_GLOBAL#northernmost_latitude=90', 'NC_GLOBAL#number_of_columns=8640', 'NC_GLOBAL#number_of_lines=4320', 'NC_GLOBAL#platform=Aqua', 'NC_GLOBAL#processing_level=L3 Mapped', 'NC_GLOBAL#processing_version=2018.0', 'NC_GLOBAL#project=Ocean Biology Processing Group (NASA/GSFC/OBPG)', 'NC_GLOBAL#publisher_email=data@oceancolor.gsfc.nasa.gov', 'NC_GLOBAL#publisher_name=NASA/GSFC/OBPG', 'NC_GLOBAL#publisher_url=http://oceandata.sci.gsfc.nasa.gov', 'NC_GLOBAL#southernmost_latitude=-90', 'NC_GLOBAL#spatialResolution=4.64 km', 'NC_GLOBAL#standard_name_vocabulary=CF Standard Name Table v36', 'NC_GLOBAL#start_orbit_number=88173', 'NC_GLOBAL#suggested_image_scaling_applied=No', 'NC_GLOBAL#suggested_image_scaling_maximum=20', 'NC_GLOBAL#suggested_image_scaling_minimum=0.0099999998', 'NC_GLOBAL#suggested_image_scaling_type=LOG', 'NC_GLOBAL#sw_point_latitude=-89.979164', 'NC_GLOBAL#sw_point_longitude=-179.97917', 'NC_GLOBAL#temporal_range=month', 'NC_GLOBAL#title=MODISA Level-3 Standard Mapped Image', 'NC_GLOBAL#westernmost_longitude=-180']
license: {'documentationUrl': '', 'dataProviderName': 'NASA', 'dataProviderUrl': '', 'licenseId': '', 'dataPolicy': '', 'doi': '', 'credits': ''}
maxValue: [99.99925994873]
minValue: [0.00099999993108213]
noDataValue: -32767
numberOfRecords: 183
profile: {'profileSchema': 'eo_profile_schema.json', 'name': 'Earth Observation', 'mission': 'MODIS', 'sensor': 'MODIS', 'processingLevel': 'L3 Mapped', 'instrument': 'MODIS', 'platform': 'MOD/MYD'}
resolutionUnit: degree
temporalResolution: Monthly
unit: mg m^-3
unitDescription: Temperature
updateDate: 2021-07-09T14:47:36Z
geometry: {'type': 'Polygon', 'coordinates': [[[-180.0, -90.0], [180.0, -90.0], [180.0, 89.9999975], [-180.0, 89.9999975], [-180.0, -90.0]]]}
resolutions: [0.0416666678441787]
anyText: 4326,NASA,Daily,NASA
applications: ['Marine']
datasetManager: govoni@meeo.it
datasetManagerOrganisation: meeo
description: MODIS Chlorophyll-a Concentration
This algorithm returns the near-surface concentration of chlorophyll-a (chlor_a) in mg m-3, calculated using an empirical relationship derived from in situ measurements of chlor_a and remote sensing reflectances (Rrs) in the blue-to-green region of the visible spectrum. The implementation is contingent on the availability three or more sensor bands spanning the 440 - 670 nm spectral regime. The algorithm is applicable to all current ocean color sensors. The chlor_a product is included as part of the standard Level-2 OC product suite and the Level-3 CHL product suite.
The current implementation for the default chlorophyll algorithm (chlor_a) employs the standard OC3/OC4 (OCx) band ratio algorithm merged with the color index (CI) of Hu et al. (2012). As described in that paper, this refinement is restricted to relatively clear water, and the general impact is to reduce artifacts and biases in clear-water chlorophyll retrievals due to residual glint, stray light, atmospheric correction errors, and white or spectrally-linear bias errors in Rrs. As implemented, the algorithm diverges slightly from what was published in Hu et al. (2012) in that the transition between CI and OCx now occurs at 0.15 < CI < 0.2 mg/m3 to ensure a smooth transition.
title: Ocean Color Data: Modis-aqua_chl-a (monthly)
timeReferenceSystem: UTC
units: mg m^-3
unitsDescription: Chlorophyll Concentration
services: ['MWCS', 'ADAM', 'AdamApi']
technicalManager: govoni@meeo.it
filtersEnabled: {'type': 'object', 'title': 'Dataset filters', 'properties': {'startDate': {'title': 'Start Date', 'type': 'string', 'format': 'date', 'text_rule': 'false', 'ops_only': 'false', 'math_rule': 'false', 'order_rule': 'false', 'pattern': '^d{4}-[01]d-[0-3]d(T[0-2]d:[0-9]d:[0-9]dZ)?$'}, 'endDate': {'title': 'End Date', 'type': 'string', 'format': 'date', 'ops_only': 'false', 'text_rule': 'false', 'math_rule': 'false', 'order_rule': 'false', 'pattern': '^d{4}-[01]d-[0-3]d(T[0-2]d:[0-9]d:[0-9]dZ)?$'}}}
starDate: 2002-07-04T00:40:05Z
endDate: 2018-12-01T00:10:01Z
accounting: {'unit': 0.0032}
Discover and select products from a dataset¶
for a given time range and spatial coverage
Get data over Artic region¶
The geometry field is extracted from a GeoJSON object , retrieving the value of the “feature” element.
Search data¶
only print the first 10 products
from adamapi import Search
The GeoJson object needs to be rearranged according to the counterclockwise winding order.This operation is executed in the next few lines to obtain a geometry that meets the requirements of the method. Geom_1 is the final result to be used in the discovery operation.
geom_1 = "{'type': 'Polygon', 'coordinates': [[[-180, 90], [180, 90], [180, 60], [-180, 60], [-180, 90]]]}"
start_date = '2003-01-01'
end_date = '2003-12-31'
search = Search( a )
results = search.getProducts(
datasetID,
geometry= geom_1,
startDate = start_date,
endDate = end_date
)
# Printing the results
print('\033[1m' + 'List of available products:')
print ('\033[0m')
count = 1
for i in results['content']:
print("\033[1;31;1m" + "#" + str(count))
print ('\033[0m')
for k in i.items():
print(str(k[0]) + ': ' + str(k[1]))
count = count+1
print('------------------------------------')
List of available products:
#1
_id: {'$oid': '60e85ffc6dfebc0806d0ec1e'}
datasetId: MODh20chlMO_4km
productDate: 2003-12-01T00:05:00Z
productId: NETCDF:"/mnt/VOL100T/reliance/MODISA_L3m_CHL/2003/12/01/A20033352003365.L3m_MO_CHL_chlor_a_4km.nc":chlor_a
band: 1
bandDescription: mass_concentration_chlorophyll_concentration_in_sea_water
geometry: {'geometries': [{'type': 'Polygon', 'coordinates': [[[-180, -90], [0.0, -90], [0.0, 89.9999975], [-180, 89.9999975], [-180, -90]]]}, {'type': 'Polygon', 'coordinates': [[[0.0, -90], [180, -90], [180, 89.9999975], [0.0, 89.9999975], [0.0, -90]]]}], 'type': 'GeometryCollection'}
insertDate: 2021-07-09T14:41:00Z
platform: A20
status: Online
------------------------------------
#2
_id: {'$oid': '60e860146dfebc0806d0f073'}
datasetId: MODh20chlMO_4km
productDate: 2003-11-01T00:00:00Z
productId: NETCDF:"/mnt/VOL100T/reliance/MODISA_L3m_CHL/2003/11/01/A20033052003334.L3m_MO_CHL_chlor_a_4km.nc":chlor_a
band: 1
bandDescription: mass_concentration_chlorophyll_concentration_in_sea_water
geometry: {'geometries': [{'type': 'Polygon', 'coordinates': [[[-180, -90], [0.0, -90], [0.0, 89.9999975], [-180, 89.9999975], [-180, -90]]]}, {'type': 'Polygon', 'coordinates': [[[0.0, -90], [180, -90], [180, 89.9999975], [0.0, 89.9999975], [0.0, -90]]]}], 'type': 'GeometryCollection'}
insertDate: 2021-07-09T14:41:23Z
platform: A20
status: Online
------------------------------------
#3
_id: {'$oid': '60e8602c6dfebc0806d0fbe4'}
datasetId: MODh20chlMO_4km
productDate: 2003-10-01T00:30:01Z
productId: NETCDF:"/mnt/VOL100T/reliance/MODISA_L3m_CHL/2003/10/01/A20032742003304.L3m_MO_CHL_chlor_a_4km.nc":chlor_a
band: 1
bandDescription: mass_concentration_of_chlorophyll_in_sea_water
geometry: {'geometries': [{'type': 'Polygon', 'coordinates': [[[-180, -90], [0.0, -90], [0.0, 89.9999975], [-180, 89.9999975], [-180, -90]]]}, {'type': 'Polygon', 'coordinates': [[[0.0, -90], [180, -90], [180, 89.9999975], [0.0, 89.9999975], [0.0, -90]]]}], 'type': 'GeometryCollection'}
insertDate: 2021-07-09T14:41:48Z
platform: A20
status: Online
------------------------------------
#4
_id: {'$oid': '60e860436dfebc0806d10158'}
datasetId: MODh20chlMO_4km
productDate: 2003-09-01T00:20:01Z
productId: NETCDF:"/mnt/VOL100T/reliance/MODISA_L3m_CHL/2003/09/01/A20032442003273.L3m_MO_CHL_chlor_a_4km.nc":chlor_a
band: 1
bandDescription: mass_concentration_chlorophyll_concentration_in_sea_water
geometry: {'geometries': [{'type': 'Polygon', 'coordinates': [[[-180, -90], [0.0, -90], [0.0, 89.9999975], [-180, 89.9999975], [-180, -90]]]}, {'type': 'Polygon', 'coordinates': [[[0.0, -90], [180, -90], [180, 89.9999975], [0.0, 89.9999975], [0.0, -90]]]}], 'type': 'GeometryCollection'}
insertDate: 2021-07-09T14:42:11Z
platform: A20
status: Online
------------------------------------
#5
_id: {'$oid': '60e8605b6dfebc0806d106b6'}
datasetId: MODh20chlMO_4km
productDate: 2003-08-01T00:50:00Z
productId: NETCDF:"/mnt/VOL100T/reliance/MODISA_L3m_CHL/2003/08/01/A20032132003243.L3m_MO_CHL_chlor_a_4km.nc":chlor_a
band: 1
bandDescription: mass_concentration_chlorophyll_concentration_in_sea_water
geometry: {'geometries': [{'type': 'Polygon', 'coordinates': [[[-180, -90], [0.0, -90], [0.0, 89.9999975], [-180, 89.9999975], [-180, -90]]]}, {'type': 'Polygon', 'coordinates': [[[0.0, -90], [180, -90], [180, 89.9999975], [0.0, 89.9999975], [0.0, -90]]]}], 'type': 'GeometryCollection'}
insertDate: 2021-07-09T14:42:35Z
platform: A20
status: Online
------------------------------------
#6
_id: {'$oid': '60e860746dfebc0806d10d6c'}
datasetId: MODh20chlMO_4km
productDate: 2003-07-01T00:10:01Z
productId: NETCDF:"/mnt/VOL100T/reliance/MODISA_L3m_CHL/2003/07/01/A20031822003212.L3m_MO_CHL_chlor_a_4km.nc":chlor_a
band: 1
bandDescription: mass_concentration_chlorophyll_concentration_in_sea_water
geometry: {'geometries': [{'type': 'Polygon', 'coordinates': [[[-180, -90], [0.0, -90], [0.0, 89.9999975], [-180, 89.9999975], [-180, -90]]]}, {'type': 'Polygon', 'coordinates': [[[0.0, -90], [180, -90], [180, 89.9999975], [0.0, 89.9999975], [0.0, -90]]]}], 'type': 'GeometryCollection'}
insertDate: 2021-07-09T14:43:00Z
platform: A20
status: Online
------------------------------------
#7
_id: {'$oid': '60e8608b6dfebc0806d11551'}
datasetId: MODh20chlMO_4km
productDate: 2003-06-01T00:00:00Z
productId: NETCDF:"/mnt/VOL100T/reliance/MODISA_L3m_CHL/2003/06/01/A20031522003181.L3m_MO_CHL_chlor_a_4km.nc":chlor_a
band: 1
bandDescription: mass_concentration_chlorophyll_concentration_in_sea_water
geometry: {'geometries': [{'type': 'Polygon', 'coordinates': [[[-180, -90], [0.0, -90], [0.0, 89.9999975], [-180, 89.9999975], [-180, -90]]]}, {'type': 'Polygon', 'coordinates': [[[0.0, -90], [180, -90], [180, 89.9999975], [0.0, 89.9999975], [0.0, -90]]]}], 'type': 'GeometryCollection'}
insertDate: 2021-07-09T14:43:22Z
platform: A20
status: Online
------------------------------------
#8
_id: {'$oid': '60e860a26dfebc0806d11582'}
datasetId: MODh20chlMO_4km
productDate: 2003-05-01T00:35:00Z
productId: NETCDF:"/mnt/VOL100T/reliance/MODISA_L3m_CHL/2003/05/01/A20031212003151.L3m_MO_CHL_chlor_a_4km.nc":chlor_a
band: 1
bandDescription: mass_concentration_chlorophyll_concentration_in_sea_water
geometry: {'geometries': [{'type': 'Polygon', 'coordinates': [[[-180, -90], [0.0, -90], [0.0, 89.9999975], [-180, 89.9999975], [-180, -90]]]}, {'type': 'Polygon', 'coordinates': [[[0.0, -90], [180, -90], [180, 89.9999975], [0.0, 89.9999975], [0.0, -90]]]}], 'type': 'GeometryCollection'}
insertDate: 2021-07-09T14:43:45Z
platform: A20
status: Online
------------------------------------
#9
_id: {'$oid': '60e860b96dfebc0806d1193c'}
datasetId: MODh20chlMO_4km
productDate: 2003-04-01T00:25:01Z
productId: NETCDF:"/mnt/VOL100T/reliance/MODISA_L3m_CHL/2003/04/01/A20030912003120.L3m_MO_CHL_chlor_a_4km.nc":chlor_a
band: 1
bandDescription: mass_concentration_chlorophyll_concentration_in_sea_water
geometry: {'geometries': [{'type': 'Polygon', 'coordinates': [[[-180, -90], [0.0, -90], [0.0, 89.9999975], [-180, 89.9999975], [-180, -90]]]}, {'type': 'Polygon', 'coordinates': [[[0.0, -90], [180, -90], [180, 89.9999975], [0.0, 89.9999975], [0.0, -90]]]}], 'type': 'GeometryCollection'}
insertDate: 2021-07-09T14:44:09Z
platform: A20
status: Online
------------------------------------
#10
_id: {'$oid': '60e860d16dfebc0806d11e30'}
datasetId: MODh20chlMO_4km
productDate: 2003-03-01T00:35:00Z
productId: NETCDF:"/mnt/VOL100T/reliance/MODISA_L3m_CHL/2003/03/01/A20030602003090.L3m_MO_CHL_chlor_a_4km.nc":chlor_a
band: 1
bandDescription: mass_concentration_chlorophyll_concentration_in_sea_water
geometry: {'geometries': [{'type': 'Polygon', 'coordinates': [[[-180, -90], [0.0, -90], [0.0, 89.9999975], [-180, 89.9999975], [-180, -90]]]}, {'type': 'Polygon', 'coordinates': [[[0.0, -90], [180, -90], [180, 89.9999975], [0.0, 89.9999975], [0.0, -90]]]}], 'type': 'GeometryCollection'}
insertDate: 2021-07-09T14:44:33Z
platform: A20
status: Online
------------------------------------
Get data¶
be aware that you alwasy get daily average from ADAM-API
def getZipData(auth, dataset_info):
if not (pathlib.Path(pathlib.Path(dataset_info['outputFname']).stem).exists() or pathlib.Path(dataset_info['outputFname']).exists()):
data = adam.GetData(auth)
image = data.getData(
datasetId = dataset_info['datasetID'],
startDate = dataset_info['startDate'],
endDate = dataset_info['endDate'],
geometry = dataset_info['geometry'],
outputFname = dataset_info['outputFname'])
print(image)
%%time
output_file = './MOD_Aqua_mass_concentration_chlorophyll_concentration_in_sea_water_' + start_date + '-' + end_date + '.zip'
datasetInfo = {
'datasetID' : datasetID,
'startDate' : start_date,
'endDate' : end_date,
'geometry' : geom_1,
'outputFname' : output_file
}
getZipData(a, datasetInfo)
./MOD_Aqua_mass_concentration_chlorophyll_concentration_in_sea_water_2003-01-01-2003-12-31.zip
CPU times: user 4.15 s, sys: 148 ms, total: 4.29 s
Wall time: 1min 7s
Data analysis and Visualization¶
Unzip data¶
def unzipData(filename):
with zipfile.ZipFile(filename, 'r') as zip_ref:
zip_ref.extractall(path = pathlib.Path(filename).stem)
if not pathlib.Path(pathlib.Path(output_file).stem).exists():
unzipData(output_file)
Read data in xarray¶
def paths_to_datetimeindex(paths):
return [datetime.strptime(date.split('_')[-1].split('.')[0], '%Y-%m-%dt%f') for date in paths]
def getData(dirtif, varname):
geotiff_list = glob.glob(dirtif)
# Create variable used for time axis
time_var = xr.Variable('time', paths_to_datetimeindex(geotiff_list))
# Load in and concatenate all individual GeoTIFFs
geotiffs_da = xr.concat([xr.open_rasterio(i, parse_coordinates=True) for i in geotiff_list],
dim=time_var)
# Covert our xarray.DataArray into a xarray.Dataset
geotiffs_da = geotiffs_da.to_dataset('band')
# Rename the dimensions to make it CF-convention compliant
geotiffs_da = geotiffs_da.rename_dims({'y': 'latitude', 'x':'longitude'})
# Rename the variable to a more useful name
geotiffs_da = geotiffs_da.rename_vars({1: varname, 'y':'latitude', 'x':'longitude'})
# set attribute to variable
geotiffs_da[varname].attrs = {'units' : geotiffs_da.attrs[varname + '#units'], 'long_name' : geotiffs_da.attrs[varname + '#long_name']}
return geotiffs_da
path_files = os.path.join(pathlib.Path(output_file).stem, '*.tif')
geotiff_ds = getData(path_files, 'chlor_a')
/data/jwd/main/035/876/35876205/tmp/ipykernel_5020/2431546948.py:6: DeprecationWarning: open_rasterio is Deprecated in favor of rioxarray. For information about transitioning, see: https://corteva.github.io/rioxarray/stable/getting_started/getting_started.html
geotiffs_da = xr.concat([xr.open_rasterio(i, parse_coordinates=True) for i in geotiff_list],
/data/jwd/main/035/876/35876205/tmp/ipykernel_5020/2431546948.py:6: DeprecationWarning: open_rasterio is Deprecated in favor of rioxarray. For information about transitioning, see: https://corteva.github.io/rioxarray/stable/getting_started/getting_started.html
geotiffs_da = xr.concat([xr.open_rasterio(i, parse_coordinates=True) for i in geotiff_list],
%%time
geotiff_ds.load()
<xarray.Dataset>
Dimensions: (time: 11, latitude: 720, longitude: 8640)
Coordinates:
* latitude (latitude) float64 89.98 89.94 89.9 89.85 ... 60.1 60.06 60.02
* longitude (longitude) float64 -180.0 -179.9 -179.9 ... 179.9 179.9 180.0
* time (time) datetime64[ns] 2003-02-01 2003-03-01 ... 2003-12-01
Data variables:
chlor_a (time, latitude, longitude) float32 -3.277e+04 ... -3.277e+04
Attributes: (12/128)
transform: (0.0416...
crs: +init=e...
res: (0.0416...
is_tiled: 0
nodatavals: (-32767...
scales: (1.0,)
... ...
NC_GLOBAL#temporal_range: month
NC_GLOBAL#time_coverage_end: 2003-03...
NC_GLOBAL#time_coverage_start: 2003-02...
NC_GLOBAL#title: HMODISA...
NC_GLOBAL#westernmost_longitude: -180
NC_GLOBAL#_lastModified: 2018-01...- time: 11
- latitude: 720
- longitude: 8640
- latitude(latitude)float6489.98 89.94 89.9 ... 60.06 60.02
array([89.979164, 89.937497, 89.895831, ..., 60.104162, 60.062496, 60.020829])
- longitude(longitude)float64-180.0 -179.9 ... 179.9 180.0
array([-179.979172, -179.937505, -179.895838, ..., 179.895838, 179.937505, 179.979172]) - time(time)datetime64[ns]2003-02-01 ... 2003-12-01
array(['2003-02-01T00:00:00.000000000', '2003-03-01T00:00:00.000000000', '2003-04-01T00:00:00.000000000', '2003-05-01T00:00:00.000000000', '2003-06-01T00:00:00.000000000', '2003-07-01T00:00:00.000000000', '2003-08-01T00:00:00.000000000', '2003-09-01T00:00:00.000000000', '2003-10-01T00:00:00.000000000', '2003-11-01T00:00:00.000000000', '2003-12-01T00:00:00.000000000'], dtype='datetime64[ns]')
- chlor_a(time, latitude, longitude)float32-3.277e+04 ... -3.277e+04
- units :
- mg m^-3
- long_name :
- Chlorophyll Concentration, OCI Algorithm
array([[[-3.2767000e+04, -3.2767000e+04, -3.2767000e+04, ..., -3.2767000e+04, -3.2767000e+04, -3.2767000e+04], [-3.2767000e+04, -3.2767000e+04, -3.2767000e+04, ..., -3.2767000e+04, -3.2767000e+04, -3.2767000e+04], [-3.2767000e+04, -3.2767000e+04, -3.2767000e+04, ..., -3.2767000e+04, -3.2767000e+04, -3.2767000e+04], ..., [-3.2767000e+04, -3.2767000e+04, -3.2767000e+04, ..., -3.2767000e+04, -3.2767000e+04, -3.2767000e+04], [-3.2767000e+04, -3.2767000e+04, -3.2767000e+04, ..., -3.2767000e+04, -3.2767000e+04, -3.2767000e+04], [-3.2767000e+04, -3.2767000e+04, -3.2767000e+04, ..., -3.2767000e+04, -3.2767000e+04, -3.2767000e+04]], [[-3.2767000e+04, -3.2767000e+04, -3.2767000e+04, ..., -3.2767000e+04, -3.2767000e+04, -3.2767000e+04], [-3.2767000e+04, -3.2767000e+04, -3.2767000e+04, ..., -3.2767000e+04, -3.2767000e+04, -3.2767000e+04], [-3.2767000e+04, -3.2767000e+04, -3.2767000e+04, ..., -3.2767000e+04, -3.2767000e+04, -3.2767000e+04], ... [-3.2767000e+04, -3.2767000e+04, -3.2767000e+04, ..., -3.2767000e+04, -3.2767000e+04, -3.2767000e+04], [-3.2767000e+04, -3.2767000e+04, -3.2767000e+04, ..., -3.2767000e+04, -3.2767000e+04, -3.2767000e+04], [-3.2767000e+04, -3.2767000e+04, -3.2767000e+04, ..., -3.2767000e+04, -3.2767000e+04, -3.2767000e+04]], [[-3.2767000e+04, -3.2767000e+04, -3.2767000e+04, ..., -3.2767000e+04, -3.2767000e+04, -3.2767000e+04], [-3.2767000e+04, -3.2767000e+04, -3.2767000e+04, ..., -3.2767000e+04, -3.2767000e+04, -3.2767000e+04], [-3.2767000e+04, -3.2767000e+04, -3.2767000e+04, ..., -3.2767000e+04, -3.2767000e+04, -3.2767000e+04], ..., [-3.2767000e+04, -3.2767000e+04, -3.2767000e+04, ..., -3.2767000e+04, -3.2767000e+04, -3.2767000e+04], [-3.2767000e+04, -3.2767000e+04, -3.2767000e+04, ..., -3.2767000e+04, -3.2767000e+04, -3.2767000e+04], [-3.2767000e+04, -3.2767000e+04, -3.2767000e+04, ..., -3.2767000e+04, -3.2767000e+04, -3.2767000e+04]]], dtype=float32)
- transform :
- (0.0416666678441787, 0.0, -180.0000050868518, 0.0, -0.0416666690219633, 89.99999745804614)
- crs :
- +init=epsg:4326
- res :
- (0.0416666678441787, 0.0416666690219633)
- is_tiled :
- 0
- nodatavals :
- (-32767.0,)
- scales :
- (1.0,)
- offsets :
- (0.0,)
- /processing_control/input_parameters/NC_GLOBAL#alt_thresh_lat :
- 999
- /processing_control/input_parameters/NC_GLOBAL#apply_pal :
- 1
- /processing_control/input_parameters/NC_GLOBAL#central_meridian :
- -999
- /processing_control/input_parameters/NC_GLOBAL#deflate :
- 4
- /processing_control/input_parameters/NC_GLOBAL#east :
- 180.000
- /processing_control/input_parameters/NC_GLOBAL#fudge :
- 1.0
- /processing_control/input_parameters/NC_GLOBAL#ifile :
- A20030322003059.L3b_MO_CHL.nc
- /processing_control/input_parameters/NC_GLOBAL#interp :
- area
- /processing_control/input_parameters/NC_GLOBAL#land :
- $OCDATAROOT/common/landmask_GMT15ARC.nc
- /processing_control/input_parameters/NC_GLOBAL#mask_land :
- no
- /processing_control/input_parameters/NC_GLOBAL#north :
- 90.000
- /processing_control/input_parameters/NC_GLOBAL#num_cache :
- 500
- /processing_control/input_parameters/NC_GLOBAL#ofile :
- A20030322003059.L3m_MO_CHL_chlor_a_4km.nc
- /processing_control/input_parameters/NC_GLOBAL#oformat :
- 2
- /processing_control/input_parameters/NC_GLOBAL#oformat2 :
- png
- /processing_control/input_parameters/NC_GLOBAL#par :
- A20030322003059.L3m_MO_CHL_chlor_a_4km.nc.param
- /processing_control/input_parameters/NC_GLOBAL#product :
- chlor_a
- /processing_control/input_parameters/NC_GLOBAL#product_rgb :
- rhos_645,rhos_555,rhos_469
- /processing_control/input_parameters/NC_GLOBAL#projection :
- smi
- /processing_control/input_parameters/NC_GLOBAL#pversion :
- 2018.0
- /processing_control/input_parameters/NC_GLOBAL#quiet :
- false
- /processing_control/input_parameters/NC_GLOBAL#resolution :
- 4km
- /processing_control/input_parameters/NC_GLOBAL#south :
- -90.000
- /processing_control/input_parameters/NC_GLOBAL#suite :
- CHL
- /processing_control/input_parameters/NC_GLOBAL#threshold :
- 0
- /processing_control/input_parameters/NC_GLOBAL#trimNSEW :
- yes
- /processing_control/input_parameters/NC_GLOBAL#use_quality :
- yes
- /processing_control/input_parameters/NC_GLOBAL#use_rgb :
- no
- /processing_control/input_parameters/NC_GLOBAL#west :
- -180.000
- /processing_control/NC_GLOBAL#l2_flag_names :
- ATMFAIL,LAND,HILT,HISATZEN,STRAYLIGHT,CLDICE,COCCOLITH,LOWLW,CHLWARN,CHLFAIL,NAVWARN,MAXAERITER,ATMWARN,HISOLZEN,NAVFAIL,FILTER,HIGLINT
- /processing_control/NC_GLOBAL#software_name :
- l3mapgen
- /processing_control/NC_GLOBAL#software_version :
- 2.0.0-V2017.0
- /processing_control/NC_GLOBAL#source :
- A20030322003059.L3b_MO_CHL.nc
- AREA_OR_POINT :
- Area
- chlor_a#display_max :
- 20
- chlor_a#display_min :
- 0.0099999998
- chlor_a#display_scale :
- log
- chlor_a#keywords :
- EARTH SCIENCE > OCEANS > OCEAN CHEMISTRY > CHLOROPHYLL
- chlor_a#long_name :
- Chlorophyll Concentration, OCI Algorithm
- chlor_a#reference :
- Hu, C., Lee Z., and Franz, B.A. (2012). Chlorophyll-a algorithms for oligotrophic oceans: A novel approach based on three-band reflectance difference, J. Geophys. Res., 117, C01011, doi:10.1029/2011JC007395.
- chlor_a#standard_name :
- mass_concentration_chlorophyll_concentration_in_sea_water
- chlor_a#units :
- mg m^-3
- chlor_a#valid_max :
- 100
- chlor_a#valid_min :
- 0.001
- chlor_a#_FillValue :
- -32767
- lat#long_name :
- Latitude
- lat#standard_name :
- latitude
- lat#units :
- degrees_north
- lat#valid_max :
- 90
- lat#valid_min :
- -90
- lat#_FillValue :
- -999
- lon#long_name :
- Longitude
- lon#standard_name :
- longitude
- lon#units :
- degrees_east
- lon#valid_max :
- 180
- lon#valid_min :
- -180
- lon#_FillValue :
- -999
- NC_GLOBAL#cdm_data_type :
- grid
- NC_GLOBAL#Conventions :
- CF-1.6 ACDD-1.3
- NC_GLOBAL#creator_email :
- data@oceancolor.gsfc.nasa.gov
- NC_GLOBAL#creator_name :
- NASA/GSFC/OBPG
- NC_GLOBAL#creator_url :
- http://oceandata.sci.gsfc.nasa.gov
- NC_GLOBAL#data_bins :
- 17780644
- NC_GLOBAL#data_maximum :
- 99.401733
- NC_GLOBAL#data_minimum :
- 0.0079426123
- NC_GLOBAL#date_created :
- 2018-01-03T23:03:57.000Z
- NC_GLOBAL#easternmost_longitude :
- 180
- NC_GLOBAL#end_orbit_number :
- 4381
- NC_GLOBAL#geospatial_lat_max :
- 90
- NC_GLOBAL#geospatial_lat_min :
- -90
- NC_GLOBAL#geospatial_lat_resolution :
- 4.6383123
- NC_GLOBAL#geospatial_lat_units :
- degrees_north
- NC_GLOBAL#geospatial_lon_max :
- 180
- NC_GLOBAL#geospatial_lon_min :
- -180
- NC_GLOBAL#geospatial_lon_resolution :
- 4.6383123
- NC_GLOBAL#geospatial_lon_units :
- degrees_east
- NC_GLOBAL#grid_mapping_name :
- latitude_longitude
- NC_GLOBAL#history :
- l3mapgen par=A20030322003059.L3m_MO_CHL_chlor_a_4km.nc.param
- NC_GLOBAL#id :
- A20030322003059.L3b_MO_CHL.nc/L3/A20030322003059.L3b_MO_CHL.nc
- NC_GLOBAL#identifier_product_doi :
- 10.5067/AQUA/MODIS/L3M/CHL/2018
- NC_GLOBAL#identifier_product_doi_authority :
- http://dx.doi.org
- NC_GLOBAL#institution :
- NASA Goddard Space Flight Center, Ocean Ecology Laboratory, Ocean Biology Processing Group
- NC_GLOBAL#instrument :
- MODIS
- NC_GLOBAL#keywords :
- Earth Science > Oceans > Ocean Chemistry > Pigments > Chlorophyll; Earth Science > Oceans > Ocean Chemistry > Chlorophyllr
- NC_GLOBAL#keywords_vocabulary :
- NASA Global Change Master Directory (GCMD) Science Keywords
- NC_GLOBAL#l2_flag_names :
- ATMFAIL,LAND,HILT,HISATZEN,STRAYLIGHT,CLDICE,COCCOLITH,LOWLW,CHLWARN,CHLFAIL,NAVWARN,MAXAERITER,ATMWARN,HISOLZEN,NAVFAIL,FILTER,HIGLINT
- NC_GLOBAL#latitude_step :
- 0.041666668
- NC_GLOBAL#latitude_units :
- degrees_north
- NC_GLOBAL#license :
- http://science.nasa.gov/earth-science/earth-science-data/data-information-policy/
- NC_GLOBAL#longitude_step :
- 0.041666668
- NC_GLOBAL#longitude_units :
- degrees_east
- NC_GLOBAL#map_projection :
- Equidistant Cylindrical
- NC_GLOBAL#measure :
- Mean
- NC_GLOBAL#naming_authority :
- gov.nasa.gsfc.sci.oceandata
- NC_GLOBAL#northernmost_latitude :
- 90
- NC_GLOBAL#number_of_columns :
- 8640
- NC_GLOBAL#number_of_lines :
- 4320
- NC_GLOBAL#platform :
- Aqua
- NC_GLOBAL#processing_level :
- L3 Mapped
- NC_GLOBAL#processing_version :
- 2018.0
- NC_GLOBAL#product_name :
- A20030322003059.L3m_MO_CHL_chlor_a_4km.nc
- NC_GLOBAL#project :
- Ocean Biology Processing Group (NASA/GSFC/OBPG)
- NC_GLOBAL#publisher_email :
- data@oceancolor.gsfc.nasa.gov
- NC_GLOBAL#publisher_name :
- NASA/GSFC/OBPG
- NC_GLOBAL#publisher_url :
- http://oceandata.sci.gsfc.nasa.gov
- NC_GLOBAL#southernmost_latitude :
- -90
- NC_GLOBAL#spatialResolution :
- 4.64 km
- NC_GLOBAL#standard_name_vocabulary :
- CF Standard Name Table v36
- NC_GLOBAL#start_orbit_number :
- 3972
- NC_GLOBAL#suggested_image_scaling_applied :
- No
- NC_GLOBAL#suggested_image_scaling_maximum :
- 20
- NC_GLOBAL#suggested_image_scaling_minimum :
- 0.0099999998
- NC_GLOBAL#suggested_image_scaling_type :
- LOG
- NC_GLOBAL#sw_point_latitude :
- -89.979164
- NC_GLOBAL#sw_point_longitude :
- -179.97917
- NC_GLOBAL#temporal_range :
- month
- NC_GLOBAL#time_coverage_end :
- 2003-03-01T02:35:00.000Z
- NC_GLOBAL#time_coverage_start :
- 2003-02-01T00:10:01.000Z
- NC_GLOBAL#title :
- HMODISA Level-3 Standard Mapped Image
- NC_GLOBAL#westernmost_longitude :
- -180
- NC_GLOBAL#_lastModified :
- 2018-01-03T23:03:57.000Z
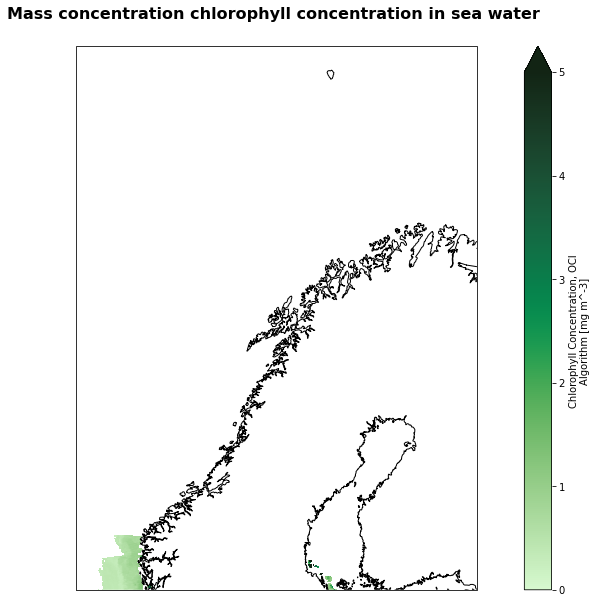
Visualization¶
%%time
extent = [0,30,60,75]
fig=plt.figure(figsize=(17,10))
# Define the projection
crs=ccrs.PlateCarree()
# We're using cartopy and are plotting in Orthographic projection
# (see documentation on cartopy)
ax = plt.subplot(1, 1, 1, projection=ccrs.Mercator(central_longitude=12.0))
ax.coastlines(resolution='10m')
ax.set_extent(extent, crs=ccrs.PlateCarree())
# We need to project our data to the new Mercator projection and for this we use `transform`.
# we set the original data projection in transform (here PlateCarree)
# we only plot values greather than 0
img = geotiff_ds['chlor_a'].isel(time=0).where(geotiff_ds['chlor_a'].isel(time=0)>0).plot(ax=ax, transform=ccrs.PlateCarree(), vmin=0, vmax=5, cmap=cmocean.cm.algae)
# Title for plot
plt.title('Mass concentration chlorophyll concentration in sea water \n',fontsize = 16, fontweight = 'bold', pad=10)
plt.savefig('Mod_Aqua-chlor_ARCTIC.png')
/srv/conda/envs/notebook/lib/python3.9/site-packages/cartopy/crs.py:825: ShapelyDeprecationWarning: __len__ for multi-part geometries is deprecated and will be removed in Shapely 2.0. Check the length of the `geoms` property instead to get the number of parts of a multi-part geometry.
if len(multi_line_string) > 1:
/srv/conda/envs/notebook/lib/python3.9/site-packages/cartopy/crs.py:877: ShapelyDeprecationWarning: Iteration over multi-part geometries is deprecated and will be removed in Shapely 2.0. Use the `geoms` property to access the constituent parts of a multi-part geometry.
for line in multi_line_string:
/srv/conda/envs/notebook/lib/python3.9/site-packages/cartopy/crs.py:944: ShapelyDeprecationWarning: __len__ for multi-part geometries is deprecated and will be removed in Shapely 2.0. Check the length of the `geoms` property instead to get the number of parts of a multi-part geometry.
if len(p_mline) > 0:
/srv/conda/envs/notebook/lib/python3.9/site-packages/cartopy/crs.py:836: ShapelyDeprecationWarning: __len__ for multi-part geometries is deprecated and will be removed in Shapely 2.0. Check the length of the `geoms` property instead to get the number of parts of a multi-part geometry.
line_strings = list(multi_line_string)
/srv/conda/envs/notebook/lib/python3.9/site-packages/cartopy/crs.py:836: ShapelyDeprecationWarning: Iteration over multi-part geometries is deprecated and will be removed in Shapely 2.0. Use the `geoms` property to access the constituent parts of a multi-part geometry.
line_strings = list(multi_line_string)
/srv/conda/envs/notebook/lib/python3.9/site-packages/cartopy/crs.py:982: ShapelyDeprecationWarning: Iteration over multi-part geometries is deprecated and will be removed in Shapely 2.0. Use the `geoms` property to access the constituent parts of a multi-part geometry.
line_strings.extend(multi_line_string)
/srv/conda/envs/notebook/lib/python3.9/site-packages/cartopy/crs.py:982: ShapelyDeprecationWarning: __len__ for multi-part geometries is deprecated and will be removed in Shapely 2.0. Check the length of the `geoms` property instead to get the number of parts of a multi-part geometry.
line_strings.extend(multi_line_string)
CPU times: user 35.2 s, sys: 515 ms, total: 35.7 s
Wall time: 36 s
geotiff_ds['chlor_a'].to_netcdf('MOD_Aqua_Chl_arctic_2003.nc')