Overview
Teaching: 20 minObjectives
- Get an overview of diagnostic package.
NorESM Diagnostic Package:
… is a NorESM model evaluation tool written with a set of scripts and utilities (bash, NCL, NCO, CDO etc) to provide a general evaluation and quick preview of the model performance with only one command line.
Components of the package:
The diagnostic tool package consists atmospheric/land components based on the NCAR package.
- CAM_DIAG: (NCAR’s AMWG Diagnostics Package)
- CLM_DIAG: (CESM Land Model Diagnostics Package)
- CICE_DIAG: snow/sea ice volume/area
- HAMOCC_DIAG: time series, climaotology, zonal mean, regional mean
- BLOM_DIAG: time series, climatologies, zonal mean, fluxes, etc
It has a one-line command interface, and is simple-to-use.
# run this wraper script without parameters shows basic usage
$ diag_run
-------------------------------------------------
Program:
/projects/NS2345K/diagnostics/noresm/bin/diag_run
Version: 2.1
-------------------------------------------------
Short description:
A wrapper script for NorESM diagnostic packages.
Basic usage:
# model-obs diagnostics
$ diag_run -m [model] -c [test case name] -s [test case start yr] -e [test case end yr]
# model1-model2 diagnostics
$ diag_run -m [model] -c [test case name] -s [test case start yr] -e [test case end yr] -c2 [cntl case name] -s2 [cntl case start yr] -e2 [cntl case end yr]
...
Two types of analysis
1. Compare model with observations
- sample plots: Historical simulation of ocean compared to observations
| 2m Air temperature | Sea surface temperature |
|---|---|
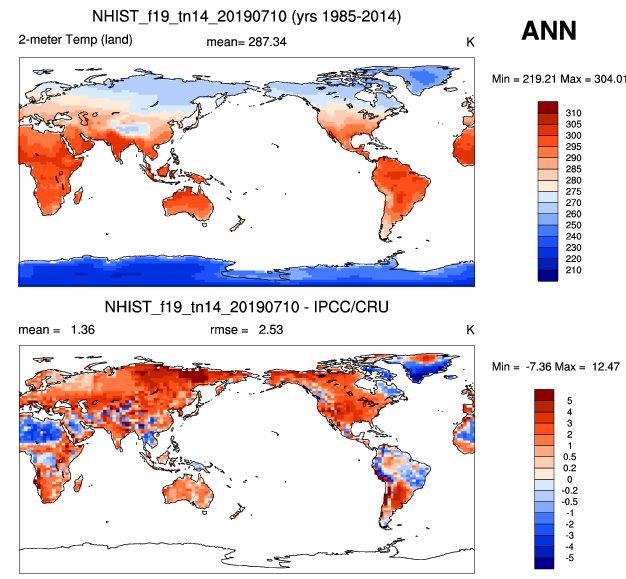 |
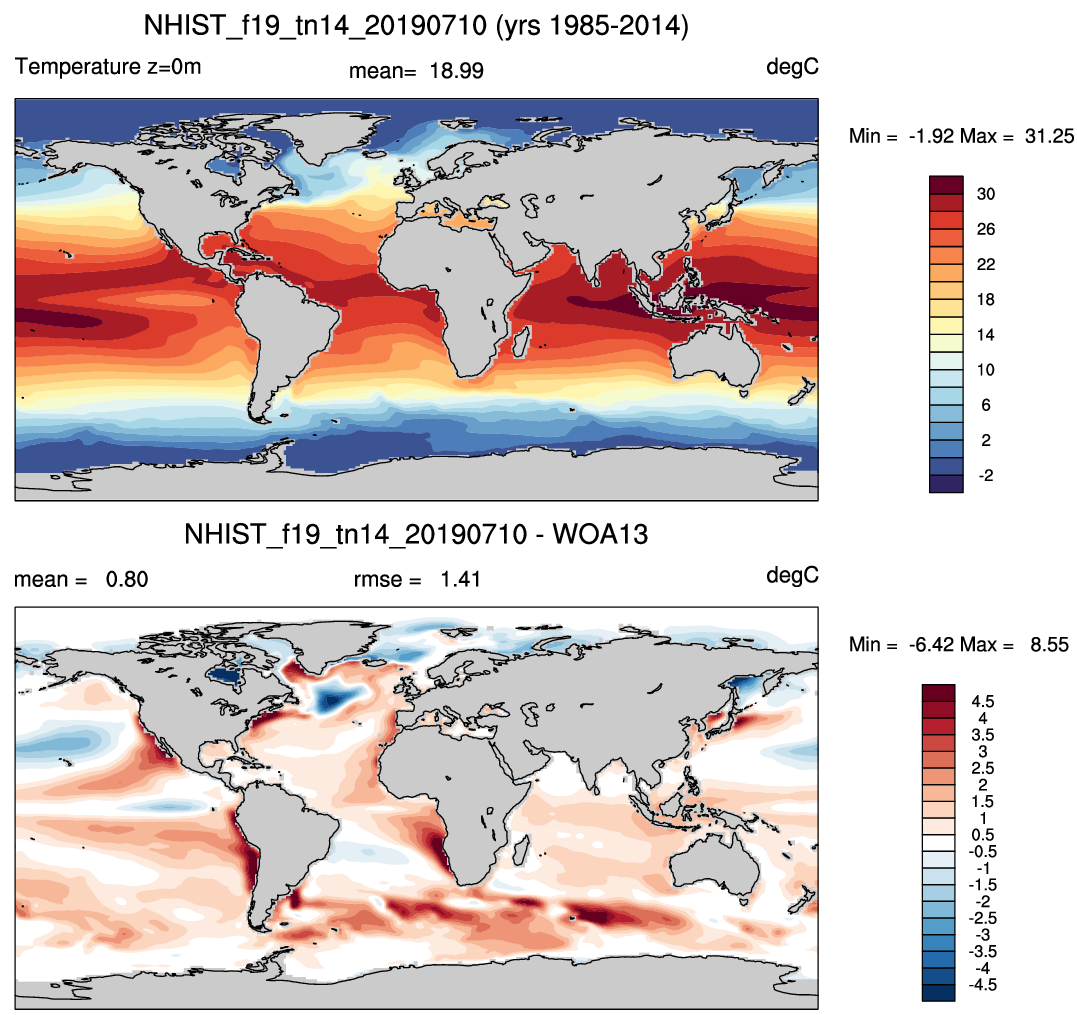 |
$ diag_run --model=cam,cice,blom \
--case=CASENAME \
--start_year=51 \
--end_year=100 \
--input-dir=/PATH/TO/MODEL/FOLDER \
--output-dir=/PATH/TO/OUTPUT/DATA \
--web-dir=/PATH/TO/GENERATED/WEBPAGES \
# or its short version
$ diag_run -m cam,cice,blom -c CASENAME -s 51 -e 100 -i /PATH/TO/MODEL/FOLDER -o /PATH/TO/OUTPUT/DATA -w /PATH/TO/GENERATED/WEBPAGES
2. Compare model with control (another simulation)
| 2m Air temperature | Sea surface temperature |
|---|---|
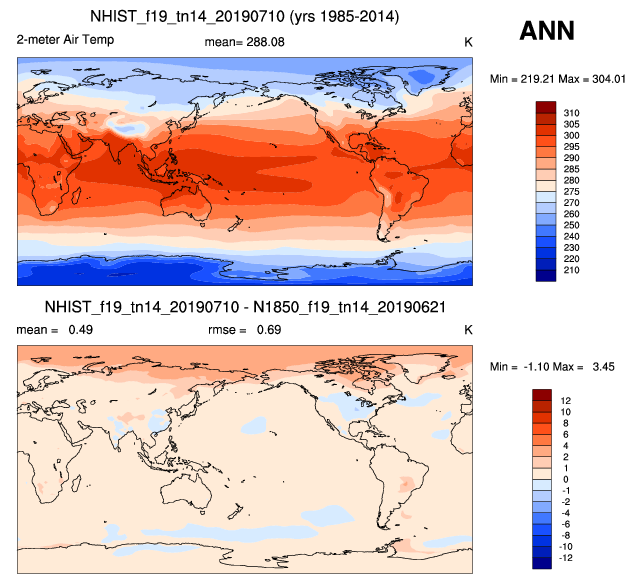 |
 |
$ diag_run --model=cam,cice,blom \
--case1=CASENAME1 \
--start_year1=51 \
--end_year1=100 \
--input-dir1=/PATH/TO/MODEL/FOLDER1 \
--case2=CASENAME2 \
--start_year2=2 \
--end_year2=50 \
--input-dir2=/PATH/TO/MODEL/FOLDER2 \
--output-dir=/PATH/TO/OUTPUT/DATA \
--web-dir=/PATH/TO/GENERATED/WEBPAGES \
# or its short version
$ diag_run -m cam,cice,blom -c1 CASENAME1 -s1 51 -e1 100 -i1 /PATH/TO/MODEL/FOLDER1 \
-c2 CASENAME2 -s2 1 -e2 50 -i2 /PATH/TO/MODEL/FOLDER2 \
-o /PATH/TO/OUTPUT/DATA \
-w /PATH/TO/GENERATED/WEBPAGES
Sets of diagnostics
| Atmospheric diagnostics (example plots) | Ocean diagnostics(Example plots) | Biogeochemistry diagnostics (Example plots) | |
|---|---|---|---|
 |
 |
 |
|
| Land diagnostics (example plots) | Sea ice diagnostics (Example plots) | ||
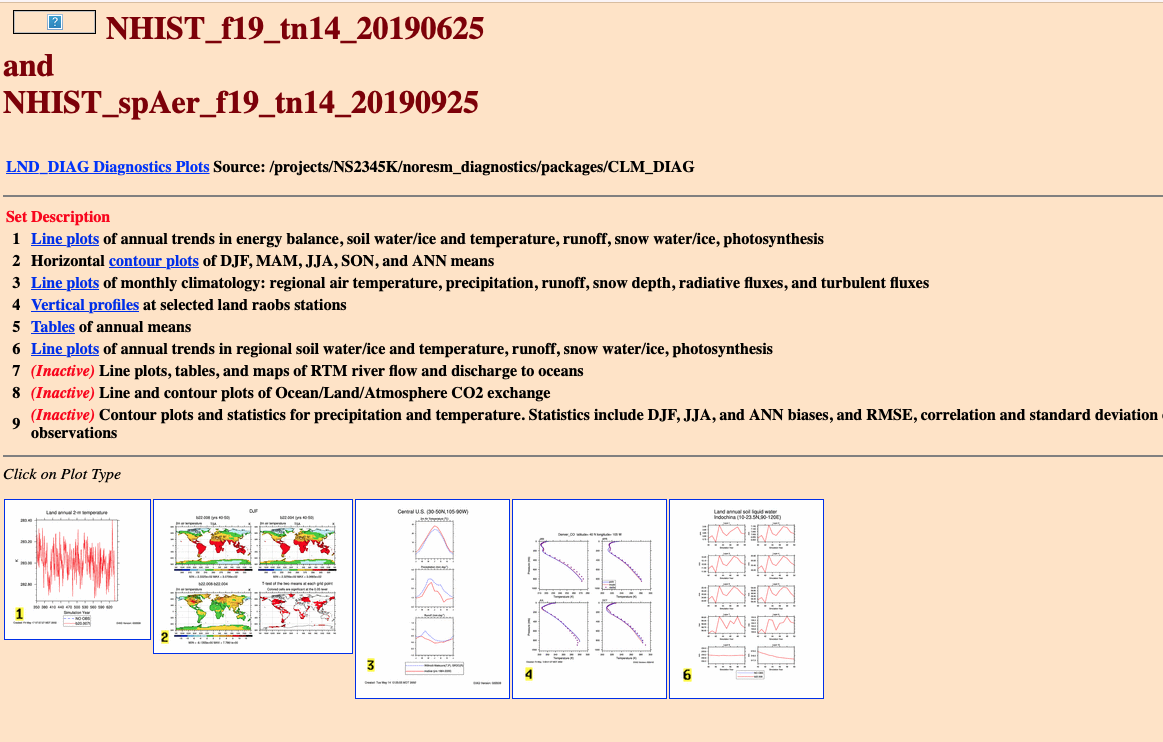 |
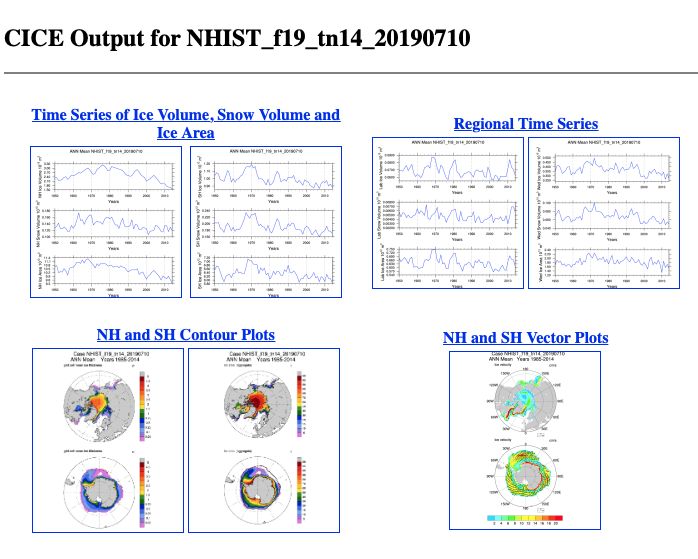 |
Browse plots online at: http://ns2345k.web.sigma2.no/diagnostics/noresm
- shared diagnostics are stored under
commom/- personal diagnostics are store under
$username/
Note
If you don’t have access to the NS2345K project, you have to specify another directory to write your webpage output by
-woption.
You can then make a tarball (tar -cvzf casenme.tar.gz /path/to/the/weboutput)
And download to your local computer to view with your browser.
Code structure
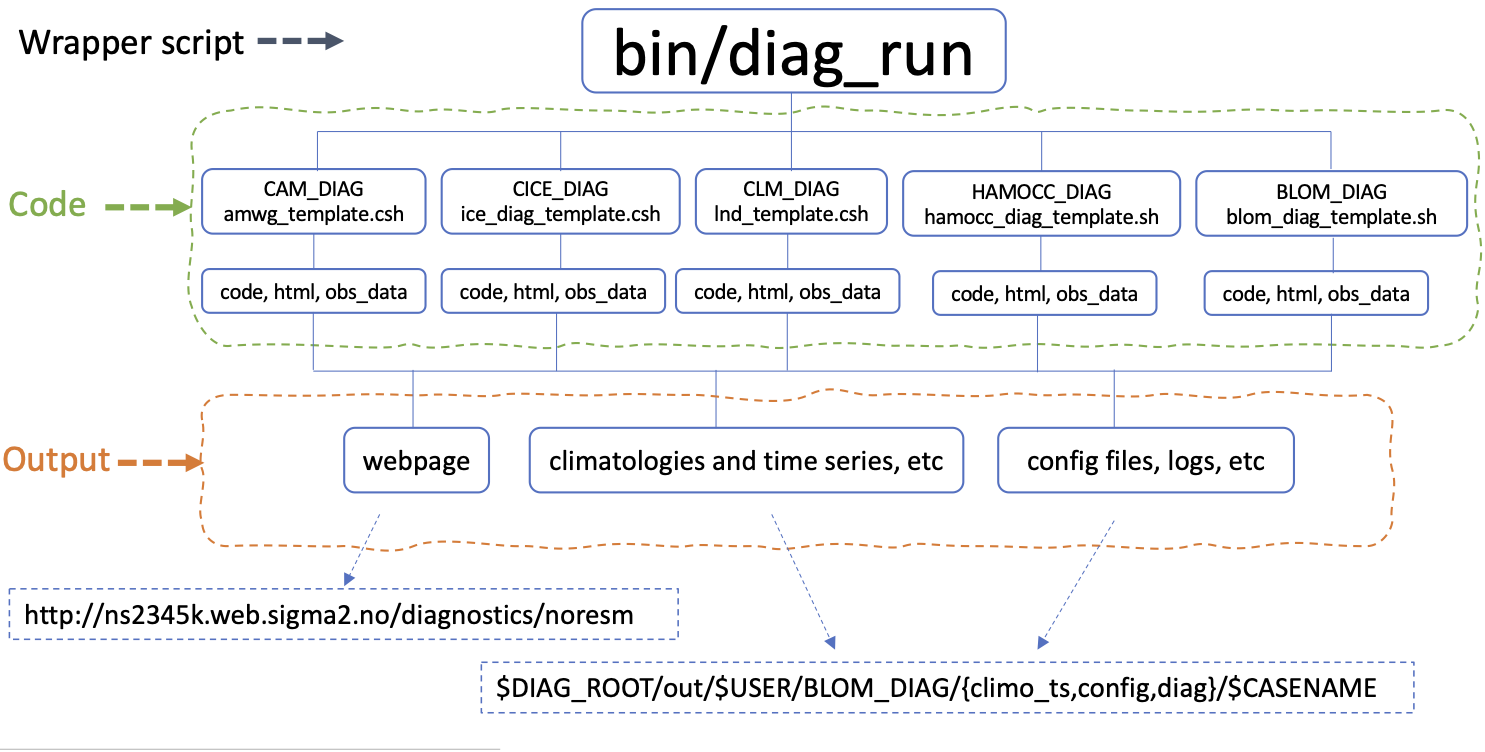
Resources
Where is it?
- Github: https://github.com/NordicESMhub/noresmdiagnostics
- NIRD: /projects/NS2345K/diagnostics/noresm
- Betzy: /trd-project1/NS2345K/diagnostics/noresm (NOT tested!)
Do NOT directly modify /trd-project1/NS2345K/diagnostics/noresm!
Install your own copy?
Yes you can, when you:
- don’t have access to NIRD at all.
- or have account on NIRD, but don’t have access to project NS2345K.
then# log on NIRD git clone https://github.com/NordicESMhub/noresmdiagnostics - or you want to make changes of the code at your will and/or want to contribute to the code.
then
You can firstforkto your private repository andcloneto install.
and then make soft links by the toolbin/linkdata.shto/projects/NS2345K/www/diagnostics/inputdata,
or download the observational data bybin/dldata.shfrom http://noresm.org/diagnostics.
Find the full doocumentation: https://noresm-docs.readthedocs.io/en/latest/diagnostics/diag_run.html
